OMIBONE: Omics-driven computer model of bone regeneration for personalized treatment
IF 3.6
2区 医学
Q2 ENDOCRINOLOGY & METABOLISM
引用次数: 0
Abstract
Treatment of bone fractures are standardized according to the AO classification, which mainly refers to the mechanical stabilization required in a given situation but neglect individual differences due to patient's healing potential or accompanying diseases. Specially in elderly or immune-compromised patients, the complexity of individual constrains on a biological as well as mechanical level are hard to account for. Here, we introduce a novel framework that allows to predict bone regeneration outcome using combined proteomic and mechanical analyses in a computer model. The framework uses Ingenuity Pathway Analysis (IPA) software to link protein changes to alterations in biological processes and integrates these in an Agent-Based Model (ABM) of bone regeneration. This combined framework allows to predict bone formation and the potential of an individual to heal a given fracture setting. The performance of the framework was evaluated by replicating the experimental setup of a mouse femur fracture stabilized with an intramedullary pin. The model was informed by serum derived proteomics data. The tissue formation patterns were compared against experimental data based on x-ray and histology images. The results indicate the framework potential in predicting an individual's bone formation potential and hold promise as a concept to enable personalized bone healing predictions for a chosen fracture fixation.
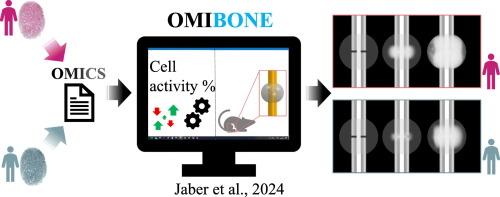
OMIBONE:用于个性化治疗的 Omics 驱动的骨再生计算机模型。
骨折的治疗是按照 AO 分类标准进行的,主要是指在特定情况下所需的机械稳定,但忽略了因患者的愈合潜力或伴随疾病而产生的个体差异。特别是对于老年人或免疫力低下的患者,很难考虑到个体在生物学和机械学层面的复杂制约因素。在这里,我们介绍了一个新颖的框架,该框架允许在计算机模型中使用蛋白质组和机械分析来预测骨再生结果。该框架使用 Ingenuity Pathway Analysis(IPA)软件将蛋白质变化与生物过程的改变联系起来,并将其整合到骨再生的代理模型(ABM)中。通过这一组合框架,可以预测骨形成情况以及个体在特定骨折情况下的愈合潜力。通过复制使用髓内针稳定小鼠股骨骨折的实验装置,对该框架的性能进行了评估。该模型参考了血清蛋白质组学数据。组织形成模式与基于 X 射线和组织学图像的实验数据进行了比较。结果表明,该框架具有预测个体骨形成潜能的潜力,并有望成为针对所选骨折固定进行个性化骨愈合预测的概念。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊
Bone
医学-内分泌学与代谢
CiteScore
8.90
自引率
4.90%
发文量
264
审稿时长
30 days
期刊介绍:
BONE is an interdisciplinary forum for the rapid publication of original articles and reviews on basic, translational, and clinical aspects of bone and mineral metabolism. The Journal also encourages submissions related to interactions of bone with other organ systems, including cartilage, endocrine, muscle, fat, neural, vascular, gastrointestinal, hematopoietic, and immune systems. Particular attention is placed on the application of experimental studies to clinical practice.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


