Triple and quadruple optimization for feature selection in cancer biomarker discovery
IF 4
2区 医学
Q2 COMPUTER SCIENCE, INTERDISCIPLINARY APPLICATIONS
引用次数: 0
Abstract
The proliferation of omics data has advanced cancer biomarker discovery but often falls short in external validation, mainly due to a narrow focus on prediction accuracy that neglects clinical utility and validation feasibility. We introduce three- and four-objective optimization strategies based on genetic algorithms to identify clinically actionable biomarkers in omics studies, addressing classification tasks aimed at distinguishing hard-to-differentiate cancer subtypes beyond histological analysis alone. Our hypothesis is that by optimizing more than one characteristic of cancer biomarkers, we may identify biomarkers that will enhance their success in external validation. Our objectives are to: (i) assess the biomarker panel’s accuracy using a machine learning (ML) framework; (ii) ensure the biomarkers exhibit significant fold-changes across subtypes, thereby boosting the success rate of PCR or immunohistochemistry validations; (iii) select a concise set of biomarkers to simplify the validation process and reduce clinical costs; and (iv) identify biomarkers crucial for predicting overall survival, which plays a significant role in determining the prognostic value of cancer subtypes. We implemented and applied triple and quadruple optimization algorithms to renal carcinoma gene expression data from TCGA. The study targets kidney cancer subtypes that are difficult to distinguish through histopathology methods. Selected RNA-seq biomarkers were assessed against the gold standard method, which relies solely on clinical information, and in external microarray-based validation datasets. Notably, these biomarkers achieved over 0.8 of accuracy in external validations and added significant value to survival predictions, outperforming the use of clinical data alone with a superior c-index. The provided tool also helps explore the trade-off between objectives, offering multiple solutions for clinical evaluation before proceeding to costly validation or clinical trials.
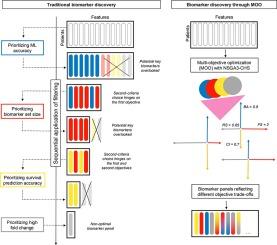
癌症生物标记物发现中特征选择的三重和四重优化。
全局组学数据的激增推动了癌症生物标记物的发现,但在外部验证方面往往存在不足,这主要是由于狭隘地关注预测准确性而忽视了临床实用性和验证可行性。我们介绍了基于遗传算法的三目标和四目标优化策略,以便在全局组学研究中发现可用于临床的生物标记物,解决旨在区分难以区分的癌症亚型的分类任务,而不仅仅是组织学分析。我们的假设是,通过优化癌症生物标志物的一个以上特征,我们可以确定生物标志物,从而提高它们在外部验证中的成功率。我们的目标是(i)使用机器学习(ML)框架评估生物标记物面板的准确性;(ii)确保生物标记物在不同亚型中表现出显著的折叠变化,从而提高 PCR 或免疫组化验证的成功率;(iii)选择一组简明的生物标记物以简化验证过程并降低临床成本;(iv)确定对预测总生存期至关重要的生物标记物,总生存期在确定癌症亚型的预后价值方面发挥着重要作用。我们对来自 TCGA 的肾癌基因表达数据实施并应用了三重和四重优化算法。这项研究的目标是组织病理学方法难以区分的肾癌亚型。对照完全依赖临床信息的金标准方法以及基于微阵列的外部验证数据集,对选定的 RNA-seq 生物标志物进行了评估。值得注意的是,这些生物标记物在外部验证中的准确率超过了 0.8,为生存预测带来了显著的价值,其 c 指数优于仅使用临床数据的方法。所提供的工具还有助于探索目标之间的权衡,在进行昂贵的验证或临床试验之前为临床评估提供多种解决方案。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Journal of Biomedical Informatics
医学-计算机:跨学科应用
CiteScore
8.90
自引率
6.70%
发文量
243
审稿时长
32 days
期刊介绍:
The Journal of Biomedical Informatics reflects a commitment to high-quality original research papers, reviews, and commentaries in the area of biomedical informatics methodology. Although we publish articles motivated by applications in the biomedical sciences (for example, clinical medicine, health care, population health, and translational bioinformatics), the journal emphasizes reports of new methodologies and techniques that have general applicability and that form the basis for the evolving science of biomedical informatics. Articles on medical devices; evaluations of implemented systems (including clinical trials of information technologies); or papers that provide insight into a biological process, a specific disease, or treatment options would generally be more suitable for publication in other venues. Papers on applications of signal processing and image analysis are often more suitable for biomedical engineering journals or other informatics journals, although we do publish papers that emphasize the information management and knowledge representation/modeling issues that arise in the storage and use of biological signals and images. System descriptions are welcome if they illustrate and substantiate the underlying methodology that is the principal focus of the report and an effort is made to address the generalizability and/or range of application of that methodology. Note also that, given the international nature of JBI, papers that deal with specific languages other than English, or with country-specific health systems or approaches, are acceptable for JBI only if they offer generalizable lessons that are relevant to the broad JBI readership, regardless of their country, language, culture, or health system.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


