Incorporation of pyridoxal-5′-phosphate into the apoenzyme: A structural study of D-amino acid transaminase from Haliscomenobacter hydrossis
IF 2.3
4区 生物学
Q3 BIOCHEMISTRY & MOLECULAR BIOLOGY
Biochimica et biophysica acta. Proteins and proteomics
Pub Date : 2024-10-13
DOI:10.1016/j.bbapap.2024.141056
引用次数: 0
Abstract
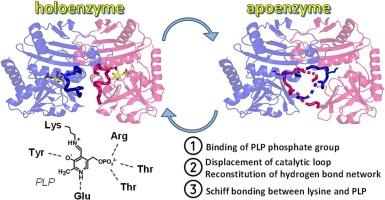
Pyridoxal-5′-phosphate (PLP)-dependent transaminases are key enzymes of amino acid metabolism in cells and remarkable biocatalysts of stereoselective amination for process chemistry applications. As cofactor-dependent enzymes, transaminases are prone to cofactor leakage. Here we discuss the holoenzyme-apoenzyme interconversion and the kinetics of PLP incorporation into the apo form of a PLP-dependent transaminase from Haliscomenobacter hydrossis. PLP binding to the apoenzyme was slow in buffer, but was accelerated in the presence of substrates. Two crystal structures of the apoenzyme were obtained: the directly obtained apoenzyme (PDB ID: 7P8O) and the one obtained by soaking crystals of the holoenzyme in a phenylhydrazine solution (PDB ID: 8YRU). The mechanism of PLP association with the apoenzyme was proposed on the basis of structural analysis of these apo forms. Three rearrangement steps, including (I) anchoring of the PLP via the phosphate group, (II) displacement of two loops, and (III) Schiff-bonding between the PLP and the ε-amino group of the catalytic lysine residue, reconstituted the active holo form of the transaminase from H. hydrossis. The results obtained allowed us to determine in the active site a permanent part and elements that are assembled by PLP, these findings may be useful for transaminase engineering for biocatalysis.
将吡哆醛-5'-磷酸掺入辅酶:水螅卤化门氏菌 D-氨基酸转氨酶的结构研究。
依赖吡哆醛-5'-磷酸(PLP)的转氨酶是细胞中氨基酸代谢的关键酶,也是工艺化学应用中立体选择性胺化的重要生物催化剂。作为依赖于辅助因子的酶,转氨酶容易发生辅助因子泄漏。在此,我们讨论了全酶与辅酶之间的相互转化,以及将 PLP 结合到 Haliscomenobacter hydrossis 的一种 PLP 依赖性转氨酶的辅酶形式中的动力学。在缓冲液中,PLP 与辅酶的结合速度较慢,但在底物存在的情况下,结合速度加快。研究人员获得了该载体酶的两种晶体结构:直接获得的载体酶(PDB ID:7P8O)和将全酶晶体浸泡在苯肼溶液中获得的晶体结构(PDB ID:8YRU)。根据对这些apo形式的结构分析,提出了PLP与apo酶结合的机制。三个重排步骤,包括(I)PLP 通过磷酸基团锚定,(II)两个环的位移,以及(III)PLP 与催化赖氨酸残基的ε-氨基之间的希夫键,重组了 H. hydrossis 的转氨酶的活性整体形式。这些结果使我们能够确定活性位点中由 PLP 组装的永久部分和元素,这些发现可能对生物催化的转氨酶工程学有用。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊
CiteScore
8.00
自引率
0.00%
发文量
55
审稿时长
33 days
期刊介绍:
BBA Proteins and Proteomics covers protein structure conformation and dynamics; protein folding; protein-ligand interactions; enzyme mechanisms, models and kinetics; protein physical properties and spectroscopy; and proteomics and bioinformatics analyses of protein structure, protein function, or protein regulation.

 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


