xU-NetFullSharp: The Novel Deep Learning Architecture for Chest X-ray Bone Shadow Suppression
IF 4.9
2区 医学
Q1 ENGINEERING, BIOMEDICAL
引用次数: 0
Abstract
Background and objectives
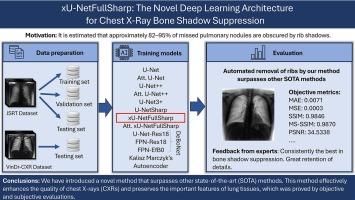
Chest X-ray image (CXR) is vital for screening, preventing, and monitoring various lung diseases. In particular, the early detection of lung cancer can significantly improve patients’ chances of survival and quality of life. Unfortunately, approximately 82–95 % of missed pulmonary nodules are estimated to be obscured by rib shadows, making them difficult to recognize. This study addresses this problem by considering the rib shadows in CXRs as noise that can be reduced using deep learning. The result of the proposed model is a CXR with improved clarity for easier and more accurate analysis by radiologists or computer algorithms.
Methods
An automated deep learning-based model for bone shadow suppression from frontal CXRs, called xU-NetFullSharp, was proposed. This network is inspired by the most modern U-NetSharp architecture and was modified using different approaches to preserve as many details as possible and accurately suppress bone shadows. For comparison, recent state-of-the-art models were implemented and trained. JSRT, VinDr-CXR, and Gusarev DES datasets were utilized for the experiments, where the JSRT dataset was extensively augmented.
Results
The performance of the proposed xU-NetFullSharp was analyzed using statistical measures and compared with that of other architectures. The proposed model significantly outperformed the others, reaching the best values of the most used metrics (0.9846 SSIM; 0.9870 MS-SSIM). It also achieves a correlation of 96.31 % and an intersection of 10.0285 between the predicted and ground truth histograms, together with the smallest value of the Bhattacharyya distance. The obtained results were validated by experts from the University Hospital Olomouc with positive feedback, thus achieving the best objective and subjective results. The proposed method has the potential to be implemented in hospital environments.
Conclusion
A comprehensive comparison of the proposed architecture with state-of-the-art methods proves its efficiency in suppressing noise in CXRs and its ability to distinguish the signals of important tissues from noise components. This methodology can potentially improve the performance of the existing CXR processing methods. The source code is released in a GitHub repository that can be accessed from the following link: https://github.com/xKev1n/xU-NetFullSharp.
xU-NetFullSharp:用于胸部 X 射线骨影抑制的新型深度学习架构
背景和目的胸部 X 光图像(CXR)对筛查、预防和监测各种肺部疾病至关重要。尤其是肺癌的早期发现可以大大提高患者的生存机会和生活质量。遗憾的是,据估计约有 82%-95% 的漏诊肺结节被肋骨阴影遮挡,因而难以辨认。为了解决这一问题,本研究将 CXR 中的肋骨阴影视为噪音,并利用深度学习降低噪音。方法提出了一种基于深度学习的自动模型,用于抑制正面 CXR 中的骨影,称为 xU-NetFullSharp。该网络受到最先进的 U-NetSharp 架构的启发,并采用不同的方法进行了修改,以保留尽可能多的细节并准确抑制骨阴影。为了进行比较,我们实施并训练了近期最先进的模型。实验使用了 JSRT、VinDr-CXR 和 Gusarev DES 数据集,其中对 JSRT 数据集进行了大量扩充。拟议模型的性能明显优于其他模型,在最常用的指标中达到了最佳值(0.9846 SSIM;0.9870 MS-SSIM)。它还实现了 96.31 % 的相关性,预测直方图与地面实况直方图之间的交集为 10.0285,巴塔查里亚距离值也最小。奥洛穆茨大学医院的专家们对所获得的结果进行了验证,并给出了积极的反馈意见,从而获得了最佳的客观和主观结果。结论 对所提出的结构与最先进的方法进行的综合比较证明,它能有效抑制 CXR 中的噪声,并能从噪声成分中区分出重要组织的信号。这种方法有可能提高现有 CXR 处理方法的性能。源代码发布在 GitHub 代码库中,可通过以下链接访问:https://github.com/xKev1n/xU-NetFullSharp。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Biomedical Signal Processing and Control
工程技术-工程:生物医学
CiteScore
9.80
自引率
13.70%
发文量
822
审稿时长
4 months
期刊介绍:
Biomedical Signal Processing and Control aims to provide a cross-disciplinary international forum for the interchange of information on research in the measurement and analysis of signals and images in clinical medicine and the biological sciences. Emphasis is placed on contributions dealing with the practical, applications-led research on the use of methods and devices in clinical diagnosis, patient monitoring and management.
Biomedical Signal Processing and Control reflects the main areas in which these methods are being used and developed at the interface of both engineering and clinical science. The scope of the journal is defined to include relevant review papers, technical notes, short communications and letters. Tutorial papers and special issues will also be published.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


