One Ring does not rule them all: Linear mtDNA in Metazoa
IF 2.6
3区 生物学
Q2 GENETICS & HEREDITY
引用次数: 0
Abstract
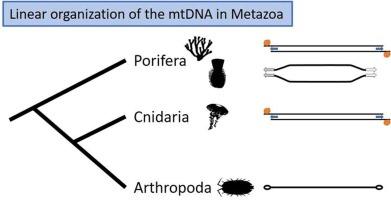
Recent advances in genome sequencing technologies have facilitated the exploration of the architecture of genomes, including mitochondrial genomes (mtDNA). In particular, whole genome sequencing has provided easier access to mitochondrial genomes with unusual organizations, which were difficult to obtain using traditional PCR-based approaches. As a consequence, there has been a steep increase in complete mtDNA sequences, particularly for Metazoa. The popular view of metazoan mtDNA is that of a small gene-dense circular chromosome. This view clashes with discoveries of a number of linear mtDNAs, particularly in non-bilaterian animals. Here, we review the distribution of linear mtDNA in Metazoa, namely in isopods, cnidarians, and sponges. We discuss the multiple origins of linear mitogenomes in these clades, where linearity has been linked to the likely insertion of a linear plasmid in cnidarians and the demosponge Acanthella acuta, while fixation of a heteroplasmy in the anticodon site of a tRNA might be responsible for the monolinear form of the mtDNA in some isopods. We also summarize our current knowledge of mechanisms that maintain the integrity of linear mitochromosomes, where a recurrent theme is the presence of terminal repeats that likely play the role of telomeres. We caution in defining a linear chromosome as complete, particularly when coding sequences and key features of linear DNA are missing. Finally, we encourage authors interested in mitogenome science to utilize all available data for linear mtDNA, including those tagged as “incomplete” or “unverified” in public databases, as they can still provide useful information such as phylogenetic characters and gene order.
一枚戒指并不能决定一切:元古宙中的线性 mtDNA
基因组测序技术的最新进展促进了对包括线粒体基因组(mtDNA)在内的基因组结构的探索。特别是,全基因组测序技术使人们更容易获得具有不寻常组织结构的线粒体基因组,而传统的基于 PCR 的方法很难获得这些基因组。因此,完整的 mtDNA 序列急剧增加,尤其是在元古宙。人们普遍认为元古宙的 mtDNA 是一个基因密集的小型环状染色体。这种观点与一些线性 mtDNA 的发现相冲突,尤其是在非两栖动物中。在这里,我们回顾了线性 mtDNA 在后生动物中的分布,即在等足类、刺丝胞动物和海绵中的分布。我们讨论了线性有丝分裂基因组在这些类群中的多种起源,在这些类群中,线性有丝分裂可能与刺胞动物和脱壳海绵 Acanthella acuta 中线性质粒的插入有关,而 tRNA 的反密码子位点的异质固定可能是一些等足类中 mtDNA 单线性形式的原因。我们还总结了我们目前对维持线粒体染色体完整性的机制的认识,其中一个经常出现的主题是末端重复序列的存在,这些重复序列可能起到端粒的作用。我们对线性染色体的完整定义持谨慎态度,尤其是当编码序列和线性 DNA 的关键特征缺失时。最后,我们鼓励对有丝分裂基因组科学感兴趣的作者利用所有可用的线性 mtDNA 数据,包括那些在公共数据库中标记为 "不完整 "或 "未经验证 "的数据,因为它们仍然可以提供有用的信息,如系统发育特征和基因顺序。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Gene
生物-遗传学
CiteScore
6.10
自引率
2.90%
发文量
718
审稿时长
42 days
期刊介绍:
Gene publishes papers that focus on the regulation, expression, function and evolution of genes in all biological contexts, including all prokaryotic and eukaryotic organisms, as well as viruses.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


