Tree smoothing: Post-hoc regularization of tree ensembles for interpretable machine learning
IF 6.8
1区 计算机科学
0 COMPUTER SCIENCE, INFORMATION SYSTEMS
引用次数: 0
Abstract
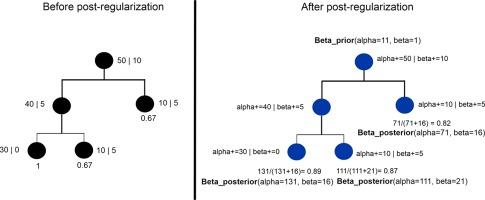
Random Forests (RFs) are powerful ensemble learning algorithms that are widely used in various machine learning tasks. However, they tend to overfit noisy or irrelevant features, which can result in decreased generalization performance. Post-hoc regularization techniques aim to solve this problem by modifying the structure of the learned ensemble after training. We propose a novel post-hoc regularization via tree smoothing for classification tasks to leverage the reliable class distributions closer to the root node whilst reducing the impact of more specific and potentially noisy splits deeper in the tree. Our novel approach allows for a form of pruning that does not alter the general structure of the trees, adjusting the influence of nodes based on their proximity to the root node. We evaluated the performance of our method on various machine learning benchmark data sets and on cancer data from The Cancer Genome Atlas (TCGA). Our approach demonstrates competitive performance compared to the state-of-the-art and, in the majority of cases, and outperforms it in most cases in terms of prediction accuracy, generalization, and interpretability.
树状平滑:为可解释的机器学习对树集合进行事后正则化
随机森林(RF)是一种强大的集合学习算法,被广泛应用于各种机器学习任务中。然而,它们往往会过度拟合噪声或不相关的特征,从而导致泛化性能下降。事后正则化技术旨在通过在训练后修改所学集合的结构来解决这一问题。我们针对分类任务提出了一种新颖的事后正则化方法,即通过树状平滑来利用靠近根节点的可靠类别分布,同时减少树状深处更具体和潜在噪声分裂的影响。我们的新方法允许在不改变树的总体结构的情况下进行剪枝,根据节点与根节点的距离调整节点的影响。我们在各种机器学习基准数据集和癌症基因组图谱(TCGA)的癌症数据上评估了我们方法的性能。与最先进的方法相比,我们的方法表现出了极具竞争力的性能,而且在大多数情况下,我们的方法在预测准确性、泛化和可解释性方面都优于最先进的方法。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Information Sciences
工程技术-计算机:信息系统
CiteScore
14.00
自引率
17.30%
发文量
1322
审稿时长
10.4 months
期刊介绍:
Informatics and Computer Science Intelligent Systems Applications is an esteemed international journal that focuses on publishing original and creative research findings in the field of information sciences. We also feature a limited number of timely tutorial and surveying contributions.
Our journal aims to cater to a diverse audience, including researchers, developers, managers, strategic planners, graduate students, and anyone interested in staying up-to-date with cutting-edge research in information science, knowledge engineering, and intelligent systems. While readers are expected to share a common interest in information science, they come from varying backgrounds such as engineering, mathematics, statistics, physics, computer science, cell biology, molecular biology, management science, cognitive science, neurobiology, behavioral sciences, and biochemistry.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


