Extracapsular extension risk assessment using an artificial intelligence prostate cancer mapping algorithm
Abstract
Objective
The objective of this study is to compare detection rates of extracapsular extension (ECE) of prostate cancer (PCa) using artificial intelligence (AI)-generated cancer maps versus MRI and conventional nomograms.
Materials and methods
We retrospectively analysed data from 147 patients who received MRI-targeted biopsy and subsequent radical prostatectomy between September 2016 and May 2022. AI-based software cleared by the United States Food and Drug Administration (Unfold AI, Avenda Health) was used to map 3D cancer probability and estimate ECE risk. Conventional ECE predictors including MRI Likert scores, capsular contact length of MRI-visible lesions, PSMA T stage, Partin tables, and the “PRedicting ExtraCapsular Extension” nomogram were used for comparison.
Postsurgical specimens were processed using whole-mount histopathology sectioning, and a genitourinary pathologist assessed each quadrant for ECE presence. ECE predictors were then evaluated on the patient (Unfold AI versus all comparators) and quadrant level (Unfold AI versus MRI Likert score). Receiver operator characteristic curves were generated and compared using DeLong's test.
Results
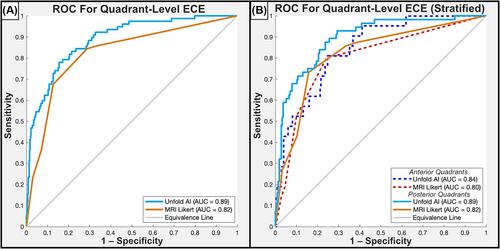
Unfold AI had a significantly higher area under the curve (AUC = 0.81) than other predictors for patient-level ECE prediction. Unfold AI achieved 68% sensitivity, 78% specificity, 71% positive predictive value, and 75% negative predictive value. At the quadrant level, Unfold AI exceeded the AUC of MRI Likert scores for posterior (0.89 versus 0.82, p = 0.003), anterior (0.84 versus 0.80, p = 0.34), and all quadrants (0.89 versus 0.82, p = 0.002). The false negative rate of Unfold AI was lower than MRI in both the anterior (−60%) and posterior prostate (−40%).
Conclusions
Unfold AI accurately predicted ECE risk, outperforming conventional methodologies. It notably improved ECE prediction over MRI in posterior quadrants, with the potential to inform nerve-spare technique and prevent positive margins. By enhancing PCa staging and risk stratification, AI-based cancer mapping may lead to better oncological and functional outcomes for patients.

 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


