Integrated analysis of transcriptome and metabolome reveals chronic low salinity stress responses in the muscle of Exopalaemon carinicauda
IF 2.2
2区 生物学
Q4 BIOCHEMISTRY & MOLECULAR BIOLOGY
Comparative Biochemistry and Physiology D-Genomics & Proteomics
Pub Date : 2024-10-11
DOI:10.1016/j.cbd.2024.101340
引用次数: 0
Abstract
Low salinity environment is one of the key factors threatening the survival of aquatic organisms. Due to the strong adaptability of low salinity, Exopalaemon carinicauda is an ideal model to study the low salinity adaptation mechanism of crustaceans. In this study, E. carinicauda from the same family were divided into two groups, which were reared at salinity of 4 ‰ and 30 ‰, respectively. Integrated analysis of transcriptome and metabolome was used to uncover the mechanisms of E. carinicauda adaptation to chronic low salinity environment. Under the chronic low salinity stress, a total of 651 differentially expressed genes (DEGs) and 386 differential metabolites (DMs) were obtained, with the majority showing downregulation. These DEGs mainly involved MAPK signal transduction pathway and structural constituent of cuticle. Besides, chitin binding and chitin metabolism process were inhibited significantly. Among the DMs, lipids and lipid-like molecules, flavor amino acids and nucleotides were detected, which may be related to the adjustment of energy metabolism and flavor of muscle. In addition, ubiquinone and other terpenoid-quinone biosynthesis pathway and alanine, aspartate, and glutamate metabolic pathway were induced. These results will enrich our understanding of the molecular mechanism underlying the chronic low salinity tolerance in E. carinicauda, providing an important theoretical basis and practical guidance for the research and breeding, thereby promoting the sustainable development of aquaculture.
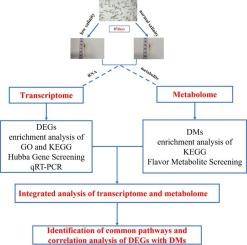
转录组和代谢组的综合分析揭示了鲤鱼肌肉中的慢性低盐度应激反应
低盐度环境是威胁水生生物生存的关键因素之一。鲤形目甲壳动物对低盐环境有较强的适应能力,是研究甲壳动物低盐适应机制的理想模型。本研究将同一科的鲤形目甲壳动物分为两组,分别在盐度为4‰和30‰的环境中饲养。通过对转录组和代谢组的综合分析,揭示了鲤鱼对慢性低盐度环境的适应机制。在慢性低盐度胁迫下,共获得了651个差异表达基因(DEGs)和386个差异代谢产物(DMs),其中大部分基因表现为下调。这些 DEGs 主要涉及 MAPK 信号转导途径和角质层结构成分。此外,几丁质结合和几丁质代谢过程也受到明显抑制。在DMs中,检测到脂质和类脂质分子、风味氨基酸和核苷酸,这可能与肌肉能量代谢和风味的调整有关。此外,还诱导了泛醌和其他萜类-醌类生物合成途径以及丙氨酸、天冬氨酸和谷氨酸代谢途径。这些结果将丰富我们对鲤鱼慢性耐低盐分子机制的认识,为研究和育种提供重要的理论依据和实践指导,从而促进水产养殖业的可持续发展。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊
CiteScore
5.10
自引率
3.30%
发文量
69
审稿时长
33 days
期刊介绍:
Comparative Biochemistry & Physiology (CBP) publishes papers in comparative, environmental and evolutionary physiology.
Part D: Genomics and Proteomics (CBPD), focuses on “omics” approaches to physiology, including comparative and functional genomics, metagenomics, transcriptomics, proteomics, metabolomics, and lipidomics. Most studies employ “omics” and/or system biology to test specific hypotheses about molecular and biochemical mechanisms underlying physiological responses to the environment. We encourage papers that address fundamental questions in comparative physiology and biochemistry rather than studies with a focus that is purely technical, methodological or descriptive in nature.

 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


