Engineering affinity of humanized ScFv targeting CD147 antibody: A combined approach of mCSM-AB2 and molecular dynamics simulations
IF 2.7
4区 生物学
Q2 BIOCHEMICAL RESEARCH METHODS
引用次数: 0
Abstract
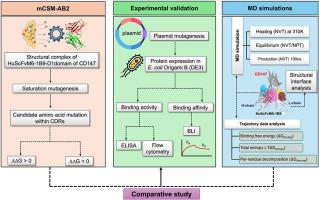
This study aims to assess the effectiveness of mCSM-AB2, a graph-based signature machine learning method, for affinity engineering of the humanized single-chain Fv anti-CD147 (HuScFvM6-1B9). In parallel, molecular dynamics (MD) simulations were used to gain valuable insights into the dynamics and affinity of the HuScFvM6-1B9-CD147 complex. The result analysis involved integrating free energy changes calculated from the mCSM-AB2 with binding free energy predictions from MD simulations. The simulated structures of the modified HuScFvM6-1B9-CD147 domain 1 complex from MD simulations were used to highlight critical residues participating in the binding surface. Interestingly, alterations in the pattern of amino acids of HuScFvM6-1B9 at the complementarity determining regions interacting with the 31EDLGS35 epitope were observed, particularly in mutants that lost binding activity. The predicted mutants of HuScFvM6-1B9 were subsequently engineered and expressed in E. coli for subsequent binding property validation. Compared to WT HuScFvM6-1B9, the mutant HuScFvM6-1B9L1:N32Y exhibited a 1.66-fold increase in binding affinity, with a KD of 1.75 × 10−8 M. While mCSM-AB2 demonstrates insignificant improvement in predicting binding affinity enhancements, it excels at predicting negative effects, aligning well with experimental validation. In addition to binding free energies, total entropy was considered to explain the discrepancy between mCSM-AB2 predictions and experimental results. This study provides guidelines and identifies the limitations of mCSM-AB2 and MD simulations in antibody engineering.
人源化 ScFv 靶向 CD147 抗体的亲和力工程:mCSM-AB2 与分子动力学模拟相结合的方法
本研究旨在评估基于图谱的特征机器学习方法 mCSM-AB2 在人源化单链 Fv 抗 CD147(HuScFvM6-1B9)亲和力工程中的有效性。与此同时,还利用分子动力学(MD)模拟来深入了解 HuScFvM6-1B9-CD147 复合物的动力学和亲和力。结果分析包括将 mCSM-AB2 计算出的自由能变化与 MD 模拟预测的结合自由能进行整合。通过 MD 模拟得出的修饰后 HuScFvM6-1B9-CD147 结构域 1 复合物的模拟结构被用来突出参与结合面的关键残基。有趣的是,在与 31EDLGS35 表位相互作用的互补性决定区域,观察到 HuScFvM6-1B9 氨基酸模式的改变,尤其是在失去结合活性的突变体中。预测的 HuScFvM6-1B9 突变体随后被设计并在大肠杆菌中表达,以进行后续的结合特性验证。与 WT HuScFvM6-1B9 相比,突变体 HuScFvM6-1B9L1:N32Y 的结合亲和力提高了 1.66 倍,KD 为 1.75 × 10-8 M。除了结合自由能之外,还考虑了总熵来解释 mCSM-AB2 预测与实验结果之间的差异。本研究为 mCSM-AB2 和 MD 模拟在抗体工程中的应用提供了指导,并指出了其局限性。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Journal of molecular graphics & modelling
生物-计算机:跨学科应用
CiteScore
5.50
自引率
6.90%
发文量
216
审稿时长
35 days
期刊介绍:
The Journal of Molecular Graphics and Modelling is devoted to the publication of papers on the uses of computers in theoretical investigations of molecular structure, function, interaction, and design. The scope of the journal includes all aspects of molecular modeling and computational chemistry, including, for instance, the study of molecular shape and properties, molecular simulations, protein and polymer engineering, drug design, materials design, structure-activity and structure-property relationships, database mining, and compound library design.
As a primary research journal, JMGM seeks to bring new knowledge to the attention of our readers. As such, submissions to the journal need to not only report results, but must draw conclusions and explore implications of the work presented. Authors are strongly encouraged to bear this in mind when preparing manuscripts. Routine applications of standard modelling approaches, providing only very limited new scientific insight, will not meet our criteria for publication. Reproducibility of reported calculations is an important issue. Wherever possible, we urge authors to enhance their papers with Supplementary Data, for example, in QSAR studies machine-readable versions of molecular datasets or in the development of new force-field parameters versions of the topology and force field parameter files. Routine applications of existing methods that do not lead to genuinely new insight will not be considered.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


