A Novel Ensemble Approach with Deep Transfer Learning for Accurate Identification of Foodborne Bacteria from Hyperspectral Microscopy
IF 2.6
4区 生物学
Q2 BIOLOGY
引用次数: 0
Abstract
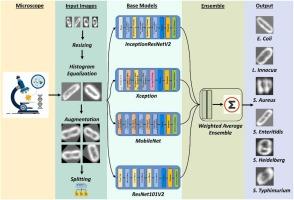
The detection of foodborne bacteria is critical in ensuring both consumer safety and food safety. If these pathogens are not properly identified, it can lead to dangerous cross-contamination. One of the most common methods for classifying bacteria is through the examination of Hyperspectral microscope imaging (HMI). A widely used technique for measuring microbial growth is microscopic cell counting. HMI is a laborious and expensive process, producing voluminous data and needing specialized equipment, which might not be widely available. Machine learning (ML) methods are now frequently utilized to automatically interpret data from hyperspectral microscopy. The objective of our study is to devise a technique that employs deep transfer learning to address the challenge of limited data and utilizes four base classifiers - InceptionResNetV2, MobileNet, ResNet101V2, and Xception - to create an ensemble-based classification model for distinguishing live and dead bacterial cells of six pathogenic strains. In order to determine the optimal weights for the base classifiers, a Powell's optimization method was utilized in conjunction with a weighted average ensemble (WAVE) technique. We carried out an extensive experimental study to verify the efficiency of our proposed ensemble model on live and dead cell images of six different foodborne bacteria. In order to gain a better understanding of the regions, we performed a Grad-CAM analysis to explain the predictions made by our model. Through a series of experiments, our proposed framework has proven its capacity to effectively and precisely detect numerous bacterial pathogens. Specifically, it achieved a perfect identification rate of 100% for Escherichia coli (EC), Listeria innocua (LI), and Salmonella Enteritidis (SE), while achieving rates of 96.30% for Salmonella Typhimurium (ST), 87.13% for Staphylococcus aureus (SA), and 94.12% for Salmonella Heidelberg (SH). As a result, it can be considered as an effective tool for the identification of foodborne pathogens, due to its high level of efficiency.
利用深度迁移学习的新型集合方法从高光谱显微镜准确识别食源性细菌
食源性细菌的检测对于确保消费者安全和食品安全至关重要。如果不能正确识别这些病原体,就会导致危险的交叉污染。对细菌进行分类的最常用方法之一是通过高光谱显微镜成像(HMI)进行检查。显微镜细胞计数是一种广泛使用的微生物生长测量技术。高光谱显微成像是一个费力且昂贵的过程,会产生大量数据,并且需要专业设备,而这些设备可能并不普及。目前,机器学习(ML)方法经常被用来自动解释高光谱显微镜的数据。我们的研究目标是设计一种技术,利用深度迁移学习来应对数据有限的挑战,并利用四个基础分类器--InceptionResNetV2、MobileNet、ResNet101V2 和 Xception--来创建一个基于集合的分类模型,以区分六种致病菌株的活细菌细胞和死细菌细胞。为了确定基础分类器的最佳权重,我们采用了鲍威尔优化法和加权平均集合(WAVE)技术。我们进行了广泛的实验研究,以验证我们提出的集合模型在六种不同食源性细菌的活细胞和死细胞图像上的效率。为了更好地了解这些区域,我们进行了 Grad-CAM 分析,以解释我们的模型所做的预测。通过一系列实验,我们提出的框架证明了其有效、精确检测多种细菌病原体的能力。具体来说,它对大肠杆菌(EC)、无毒李斯特菌(LI)和肠炎沙门氏菌(SE)的完美识别率达到了 100%,而对鼠伤寒沙门氏菌(ST)、金黄色葡萄球菌(SA)和海德堡沙门氏菌(SH)的识别率分别为 96.30%、87.13% 和 94.12%。因此,该方法因其高效率而被视为鉴定食源性病原体的有效工具。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Computational Biology and Chemistry
生物-计算机:跨学科应用
CiteScore
6.10
自引率
3.20%
发文量
142
审稿时长
24 days
期刊介绍:
Computational Biology and Chemistry publishes original research papers and review articles in all areas of computational life sciences. High quality research contributions with a major computational component in the areas of nucleic acid and protein sequence research, molecular evolution, molecular genetics (functional genomics and proteomics), theory and practice of either biology-specific or chemical-biology-specific modeling, and structural biology of nucleic acids and proteins are particularly welcome. Exceptionally high quality research work in bioinformatics, systems biology, ecology, computational pharmacology, metabolism, biomedical engineering, epidemiology, and statistical genetics will also be considered.
Given their inherent uncertainty, protein modeling and molecular docking studies should be thoroughly validated. In the absence of experimental results for validation, the use of molecular dynamics simulations along with detailed free energy calculations, for example, should be used as complementary techniques to support the major conclusions. Submissions of premature modeling exercises without additional biological insights will not be considered.
Review articles will generally be commissioned by the editors and should not be submitted to the journal without explicit invitation. However prospective authors are welcome to send a brief (one to three pages) synopsis, which will be evaluated by the editors.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


