Text mining approach for feature extraction and cartilage disease grade classification using knee MRI radiology reports
IF 4.4
2区 生物学
Q2 BIOCHEMISTRY & MOLECULAR BIOLOGY
Computational and structural biotechnology journal
Pub Date : 2024-10-05
DOI:10.1016/j.csbj.2024.10.003
引用次数: 0
Abstract
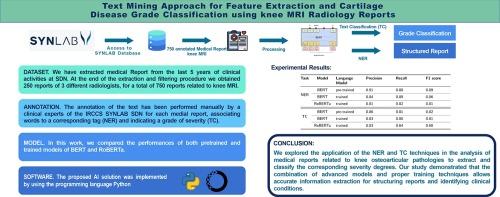
MRI radiology reporting processes can be improved by exploiting structured and semantically labelled data that can be fed to artificial intelligence (AI) tools. AI-based tools assisting radiology reporting can help to automatically individuate cartilage grading in textual magnetic resonance imaging (MRI) reports, thus supporting clinicians' decisions regarding medical imaging utilisation, diagnosis and treatment. In this study, we extracted information (clinical findings, observations, anatomical regions, etc.) and classified knee cartilage degradation from medical reports utilising transfer-learning techniques applied to the Bidirectional Encoder Representations from Transformers (BERT) model and its variants, pre-trained on an Italian-language corpus. To realise this objective, we used a dataset of 750 MRI knee reports written by three radiologists who contributed to a manual annotation process to perform text classification (TC) and named entity recognition (NER) tasks. The dataset was obtained from an internal database of the IRCCS SYNLAB SDN. Seventy percent of the dataset was used for training, 10% was used for validation and 20% was used for testing. The best-performing configurations for NER and TC tasks were based on the pre-trained BERT model. The macro F1-scores obtained with the NER and TC models are 0.89 and 0.81, respectively. The accuracies calculated on the test set for both tasks are 0.96 and 0.99, respectively.
利用膝关节磁共振成像放射学报告进行特征提取和软骨病等级分类的文本挖掘方法
核磁共振成像(MRI)放射学报告流程可通过利用结构化和语义标注数据加以改进,这些数据可输入人工智能(AI)工具。基于人工智能的放射学报告辅助工具可帮助自动对文本磁共振成像(MRI)报告中的软骨分级进行个性化处理,从而为临床医生在医学影像利用、诊断和治疗方面的决策提供支持。在这项研究中,我们从医学报告中提取信息(临床发现、观察结果、解剖区域等)并对膝关节软骨退化进行分类,将迁移学习技术应用于在意大利语语料库上预先训练的变压器双向编码器表示(BERT)模型及其变体。为实现这一目标,我们使用了由三位放射科医生撰写的 750 份核磁共振膝关节成像报告数据集,他们参与了手动注释过程,以执行文本分类(TC)和命名实体识别(NER)任务。该数据集来自 IRCCS SYNLAB SDN 的内部数据库。数据集的 70% 用于训练,10% 用于验证,20% 用于测试。NER 和 TC 任务的最佳配置基于预训练的 BERT 模型。NER 和 TC 模型获得的宏观 F1 分数分别为 0.89 和 0.81。在测试集上计算出的这两项任务的准确率分别为 0.96 和 0.99。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Computational and structural biotechnology journal
Biochemistry, Genetics and Molecular Biology-Biophysics
CiteScore
9.30
自引率
3.30%
发文量
540
审稿时长
6 weeks
期刊介绍:
Computational and Structural Biotechnology Journal (CSBJ) is an online gold open access journal publishing research articles and reviews after full peer review. All articles are published, without barriers to access, immediately upon acceptance. The journal places a strong emphasis on functional and mechanistic understanding of how molecular components in a biological process work together through the application of computational methods. Structural data may provide such insights, but they are not a pre-requisite for publication in the journal. Specific areas of interest include, but are not limited to:
Structure and function of proteins, nucleic acids and other macromolecules
Structure and function of multi-component complexes
Protein folding, processing and degradation
Enzymology
Computational and structural studies of plant systems
Microbial Informatics
Genomics
Proteomics
Metabolomics
Algorithms and Hypothesis in Bioinformatics
Mathematical and Theoretical Biology
Computational Chemistry and Drug Discovery
Microscopy and Molecular Imaging
Nanotechnology
Systems and Synthetic Biology
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


