Lessons learned from the IMMREP23 TCR-epitope prediction challenge
引用次数: 0
Abstract
Here, we present the findings from IMMREP23, the second benchmark competition focused on predicting the specificity of TCR-pMHC interactions.
The interaction of T cell receptors (TCR) towards their pMHC target is a cornerstone of the cellular immune system. Over the last decade, substantial progress has been made within the field of TCR specificity prediction, providing proof of concept for predicting TCR-pMHC interactions in a narrow space of “seen” pMHC targets where substantial training data is available. However, a significant challenge persists in extending the predictive capability to novel “unseen” pMHC targets. Furthermore, the performance of proposed methods is often challenged when evaluated outside the initial publication and data sets.
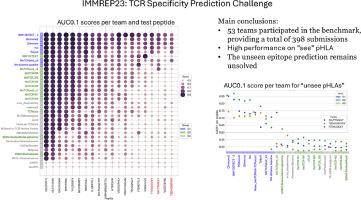
To address these issues, IMMREP23 challenge invited participants to predict, for a given test set of TCR-pMHC pairs, the likelihood that a pair would bind. A total of 53 teams participated, providing a total of 398 submissions.
The benchmark confirms that current methods achieve reasonable performance in the "seen" pMHC setting. However, most participating methods had close to random performance on the subset of “unseen” peptides, underlining that this prediction challenge remains essentially unsolved.
Finally, another key lesson from the benchmark is the critical issue of data leakage. Specifically, the data set construction procedure employed in IMMREP23 led to biases in the negative test data set. These biases were identified by several participating teams, and complicated the interpretation of the benchmark results. Based on these results, we put forward suggestions on how future competitions could avoid such data leakages and biases.
从 IMMREP23 TCR 表位预测挑战中汲取的经验教训
T 细胞受体(TCR)与其 pMHC 靶点的相互作用是细胞免疫系统的基石。在过去的十年中,TCR 特异性预测领域取得了长足的进步,证明了在有大量训练数据的情况下,在 "可见 "pMHC 靶点的狭窄空间内预测 TCR-pMHC 相互作用的概念。然而,将预测能力扩展到 "未见 "的新型 pMHC 靶点仍是一个重大挑战。为了解决这些问题,IMMREP23 挑战赛邀请参赛者针对给定的 TCR-pMHC 对测试集,预测一对 TCR-pMHC 对结合的可能性。共有 53 个团队参加,提交了 398 份报告。该基准证实,目前的方法在 "看到的 "pMHC 环境中取得了合理的性能。然而,大多数参与方法在 "未见 "肽子集上的性能接近随机,这突出表明这一预测难题基本上仍未解决。最后,基准测试的另一个关键教训是数据泄漏这一关键问题。具体来说,IMMREP23 采用的数据集构建程序导致负测试数据集出现偏差。一些参与团队发现了这些偏差,并使基准结果的解释变得复杂。基于这些结果,我们就未来的竞赛如何避免此类数据泄漏和偏差提出了建议。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Immunoinformatics (Amsterdam, Netherlands)
Immunology, Computer Science Applications
自引率
0.00%
发文量
0
审稿时长
60 days
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


