Lazy Resampling: Fast and information preserving preprocessing for deep learning
IF 4.8
2区 医学
Q1 COMPUTER SCIENCE, INTERDISCIPLINARY APPLICATIONS
引用次数: 0
Abstract
Background and Objective:
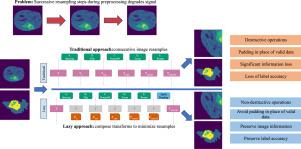
Preprocessing of data is a vital step for almost all deep learning workflows. In computer vision, manipulation of data intensity and spatial properties can improve network stability and can provide an important source of generalisation for deep neural networks. Models are frequently trained with preprocessing pipelines composed of many stages, but these pipelines come with a drawback; each stage that resamples the data costs time, degrades image quality, and adds bias to the output. Long pipelines can also be complex to design, especially in medical imaging, where cropping data early can cause significant artifacts.
Methods:
We present Lazy Resampling, a software that rephrases spatial preprocessing operations as a graphics pipeline. Rather than each transform individually modifying the data, the transforms generate transform descriptions that are composited together into a single resample operation wherever possible. This reduces pipeline execution time and, most importantly, limits signal degradation. It enables simpler pipeline design as crops and other operations become non-destructive. Lazy Resampling is designed in such a way that it provides the maximum benefit to users without requiring them to understand the underlying concepts or change the way that they build pipelines.
Results:
We evaluate Lazy Resampling by comparing traditional pipelines and the corresponding lazy resampling pipeline for the following tasks on Medical Segmentation Decathlon datasets. We demonstrate lower information loss in lazy pipelines vs. traditional pipelines. We demonstrate that Lazy Resampling can avoid catastrophic loss of semantic segmentation label accuracy occurring in traditional pipelines when passing labels through a pipeline and then back through the inverted pipeline. Finally, we demonstrate statistically significant improvements when training UNets for semantic segmentation.
Conclusion:
Lazy Resampling reduces the loss of information that occurs when running processing pipelines that traditionally have multiple resampling steps and enables researchers to build simpler pipelines by making operations such as rotation and cropping effectively non-destructive. It makes it possible to invert labels back through a pipeline without catastrophic loss of accuracy.
A reference implementation for Lazy Resampling can be found at https://github.com/KCL-BMEIS/LazyResampling. Lazy Resampling is being implemented as a core feature in MONAI, an open source python-based deep learning library for medical imaging, with a roadmap for a full integration.
懒惰重采样:用于深度学习的快速信息保护预处理
背景与目标:数据预处理是几乎所有深度学习工作流程的重要步骤。在计算机视觉领域,对数据强度和空间属性的处理可以提高网络的稳定性,并为深度神经网络提供重要的泛化来源。模型通常通过由多个阶段组成的预处理流水线进行训练,但这些流水线有一个缺点:对数据进行重新采样的每个阶段都会耗费时间、降低图像质量并增加输出偏差。长流水线的设计也很复杂,尤其是在医学成像中,早期裁剪数据会造成严重的伪影。方法:我们介绍的这款软件 "懒惰重采样"(Lazy Resampling)将空间预处理操作重新表述为图形流水线。在可能的情况下,变换生成的变换描述会被合成到一个单一的重采样操作中,而不是每个变换单独修改数据。这不仅缩短了流水线的执行时间,更重要的是限制了信号衰减。由于裁剪和其他操作都是非破坏性的,因此可以简化流水线设计。结果:我们通过比较传统管道和相应的懒惰重采样管道,对懒惰重采样进行了评估,并在医疗分割十项全能数据集上完成了以下任务。我们证明,与传统管道相比,懒惰管道的信息损失更低。我们证明了懒惰重采样可以避免传统管道在通过管道传递标签后再通过反向管道传递标签时出现的语义分割标签准确性的灾难性损失。最后,我们证明了在训练语义分割的 UNets 时在统计学上的显著改进。结论:懒惰重采样减少了在运行传统上具有多个重采样步骤的处理流水线时发生的信息损失,并通过使旋转和裁剪等操作有效地非破坏性,使研究人员能够构建更简单的流水线。懒惰重采样的参考实现可在 https://github.com/KCL-BMEIS/LazyResampling 上找到。懒惰重采样正在作为核心功能在 MONAI 中实现,MONAI 是一个基于 python 的开源深度学习库,用于医学成像,其路线图是实现全面集成。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Computer methods and programs in biomedicine
工程技术-工程:生物医学
CiteScore
12.30
自引率
6.60%
发文量
601
审稿时长
135 days
期刊介绍:
To encourage the development of formal computing methods, and their application in biomedical research and medical practice, by illustration of fundamental principles in biomedical informatics research; to stimulate basic research into application software design; to report the state of research of biomedical information processing projects; to report new computer methodologies applied in biomedical areas; the eventual distribution of demonstrable software to avoid duplication of effort; to provide a forum for discussion and improvement of existing software; to optimize contact between national organizations and regional user groups by promoting an international exchange of information on formal methods, standards and software in biomedicine.
Computer Methods and Programs in Biomedicine covers computing methodology and software systems derived from computing science for implementation in all aspects of biomedical research and medical practice. It is designed to serve: biochemists; biologists; geneticists; immunologists; neuroscientists; pharmacologists; toxicologists; clinicians; epidemiologists; psychiatrists; psychologists; cardiologists; chemists; (radio)physicists; computer scientists; programmers and systems analysts; biomedical, clinical, electrical and other engineers; teachers of medical informatics and users of educational software.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


