Evidence of RNA polymerase III recruitment and transcription at protein-coding gene promoters
IF 16.6
1区 生物学
Q1 BIOCHEMISTRY & MOLECULAR BIOLOGY
引用次数: 0
Abstract
The transcriptional interplay of human RNA polymerase I (RNA Pol I), RNA Pol II, and RNA Pol III remains largely uncharacterized due to limited integrative genomic analyses for all three enzymes. To address this gap, we applied a uniform framework to quantify global RNA Pol I, RNA Pol II, and RNA Pol III occupancies and identify both canonical and noncanonical patterns of gene localization. Most notably, our survey captures unexpected RNA Pol III recruitment at promoters of specific protein-coding genes. We show that such RNA Pol III-occupied promoters are enriched for small nascent RNAs terminating in a run of 4 Ts—a hallmark of RNA Pol III termination indicative of constrained RNA Pol III transcription. Taken further, RNA Pol III disruption generally reduces the expression of RNA Pol III-occupied protein-coding genes, suggesting RNA Pol III recruitment and transcription enhance RNA Pol II activity. These findings resemble analogous patterns of RNA Pol II activity at RNA Pol III-transcribed genes, altogether uncovering a reciprocal form of crosstalk between RNA Pol II and RNA Pol III.
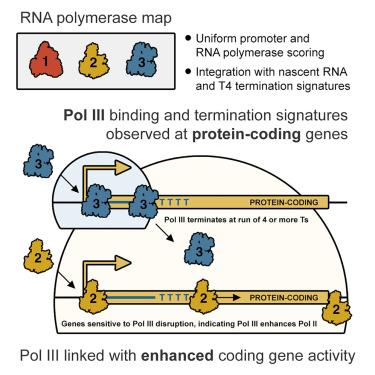
RNA 聚合酶 III 招募和转录蛋白编码基因启动子的证据
由于对人类 RNA 聚合酶 I (RNA Pol I)、RNA Pol II 和 RNA Pol III 的综合基因组分析有限,它们的转录相互作用在很大程度上仍未得到描述。为了填补这一空白,我们采用了一个统一的框架来量化全球 RNA Pol I、RNA Pol II 和 RNA Pol III 的占有率,并确定基因定位的规范和非规范模式。最值得注意的是,我们的调查捕捉到了特定蛋白编码基因启动子上意外的 RNA Pol III 招募。我们发现,这些被 RNA Pol III 占据的启动子富含以 4 个 Ts 终止的小新生 RNA--这是 RNA Pol III 终止的标志,表明 RNA Pol III 转录受限。更进一步看,RNA Pol III 的中断通常会降低 RNA Pol III 占用的编码蛋白基因的表达,这表明 RNA Pol III 的招募和转录增强了 RNA Pol II 的活性。这些发现与 RNA Pol III 转录基因上 RNA Pol II 活性的类似模式相似,共同揭示了 RNA Pol II 和 RNA Pol III 之间相互串扰的形式。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Molecular Cell
生物-生化与分子生物学
CiteScore
26.00
自引率
3.80%
发文量
389
审稿时长
1 months
期刊介绍:
Molecular Cell is a companion to Cell, the leading journal of biology and the highest-impact journal in the world. Launched in December 1997 and published monthly. Molecular Cell is dedicated to publishing cutting-edge research in molecular biology, focusing on fundamental cellular processes. The journal encompasses a wide range of topics, including DNA replication, recombination, and repair; Chromatin biology and genome organization; Transcription; RNA processing and decay; Non-coding RNA function; Translation; Protein folding, modification, and quality control; Signal transduction pathways; Cell cycle and checkpoints; Cell death; Autophagy; Metabolism.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


