A model for transcription-dependent R-loop formation at double-stranded DNA breaks: Implications for their detection and biological effects
IF 1.9
4区 数学
Q2 BIOLOGY
引用次数: 0
Abstract
R-loops are structures containing an RNA-DNA duplex and an unpaired DNA strand. During R-loop formation an RNA strand invades the DNA duplex, displacing the homologous DNA strand and binding the complementary DNA strand. Here we analyze a model for transcription-dependent R-loop formation at double-stranded DNA breaks (DSBs). In this model, R-loop formation is preceded by detachment of the non-template DNA strand from the RNA polymerase (RNAP). Then, strand exchange is initiated between the nascent RNA and the non-template DNA strand. During that strand exchange the length of the R-loop could either increase, or decrease in a biased random-walk fashion, in which the bias would depend upon the DNA sequence. Eventually, the restoration of the DNA duplex would completely displace the RNA. However, as long as the RNAP remains bound to the template DNA strand it prevents that displacement. Thus, according to the model, RNAPs stalled at DSBs can increase the lifespan of R-loops, increasing their detectability in experiments, and perhaps enhancing their biological effects.
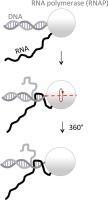
双链 DNA 断裂处转录依赖性 R 环形成模型:对其检测和生物效应的影响
R 环是包含 RNA-DNA 双链和未配对 DNA 链的结构。在 R 环形成过程中,RNA 链侵入 DNA 双链,取代同源 DNA 链并结合互补 DNA 链。在这里,我们分析了双链DNA断裂(DSB)处依赖转录的R环形成模型。在该模型中,R 环形成之前,非模板 DNA 链会从 RNA 聚合酶(RNAP)上分离。然后,新生 RNA 和非模板 DNA 链之间开始进行链交换。在链交换过程中,R 环的长度可能会增加,也可能会以有偏差的随机漫步方式减少,而偏差取决于 DNA 序列。最终,DNA 双链的恢复将完全取代 RNA。但是,只要 RNAP 仍与模板 DNA 链结合,就会阻止这种位移。因此,根据该模型,停滞在 DSB 上的 RNAP 可以延长 R 环的寿命,提高它们在实验中的可检测性,或许还能增强它们的生物效应。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊
CiteScore
4.20
自引率
5.00%
发文量
218
审稿时长
51 days
期刊介绍:
The Journal of Theoretical Biology is the leading forum for theoretical perspectives that give insight into biological processes. It covers a very wide range of topics and is of interest to biologists in many areas of research, including:
• Brain and Neuroscience
• Cancer Growth and Treatment
• Cell Biology
• Developmental Biology
• Ecology
• Evolution
• Immunology,
• Infectious and non-infectious Diseases,
• Mathematical, Computational, Biophysical and Statistical Modeling
• Microbiology, Molecular Biology, and Biochemistry
• Networks and Complex Systems
• Physiology
• Pharmacodynamics
• Animal Behavior and Game Theory
Acceptable papers are those that bear significant importance on the biology per se being presented, and not on the mathematical analysis. Papers that include some data or experimental material bearing on theory will be considered, including those that contain comparative study, statistical data analysis, mathematical proof, computer simulations, experiments, field observations, or even philosophical arguments, which are all methods to support or reject theoretical ideas. However, there should be a concerted effort to make papers intelligible to biologists in the chosen field.

 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


