Syntenic lncRNA locus exhibits DNA regulatory functions with sequence evolution
IF 2.4
3区 生物学
Q2 GENETICS & HEREDITY
引用次数: 0
Abstract
Syntenic long non-coding RNAs (lncRNAs) often show limited sequence conservation across species, prompting concern in the field. This study delves into functional signatures of syntenic lncRNAs between humans and zebrafish. Syntenic lncRNAs are highly expressed in zebrafish, with ∼90 % located near protein-coding genes, either in sense or antisense orientation. During early zebrafish development and in human embryonic stem cells (H1-hESC), syntenic lncRNA loci are enriched with cis-regulatory repressor signatures, influencing the expression of development-associated genes. In later zebrafish developmental stages and specific human cell lines, these syntenic lncRNA loci function as enhancers or transcription start sites (TSS) for protein-coding genes. Analysis of transposable elements (TEs) in syntenic lncRNA sequences revealed intriguing patterns: human lncRNAs are enriched in simple repeat elements, while their zebrafish counterparts show enrichment in LTR elements. This sequence evolution likely arises from post-rearrangement mutations that enhance DNA elements or cis-regulatory functions. It may also contribute to vertebrate innovation by creating novel transcription factor binding sites within the locus. This study highlights the conserved functionality of syntenic lncRNA loci through DNA elements, emphasizing their conserved roles across species despite sequence divergence.
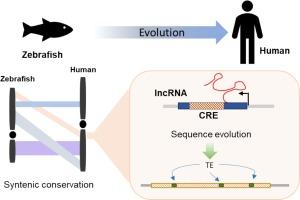
合成lncRNA基因座随着序列进化表现出DNA调控功能。
同源长非编码 RNA(lncRNA)在不同物种间往往显示出有限的序列保守性,这引起了该领域的关注。本研究深入研究了人类与斑马鱼之间的合成lncRNA的功能特征。合成lncRNA在斑马鱼中高度表达,其中90%位于蛋白编码基因附近,以有义或反义方向表达。在斑马鱼早期发育和人类胚胎干细胞(H1-hESC)中,合成lncRNA位点富含顺式调控抑制特征,影响发育相关基因的表达。在斑马鱼的后期发育阶段和特定的人类细胞系中,这些同源的 lncRNA 基因座起着蛋白编码基因的增强子或转录起始位点(TSS)的作用。对同源 lncRNA 序列中的转座元件(TEs)进行分析发现了一些有趣的模式:人类 lncRNA 富含简单重复元件,而斑马鱼的同源 lncRNA 则富含 LTR 元件。这种序列进化可能源于重组后的突变,这种突变增强了DNA元件或顺式调控功能。它还可能通过在基因座内创建新的转录因子结合位点来促进脊椎动物的创新。这项研究通过DNA元件突出了同源lncRNA基因座的保守功能,强调了尽管存在序列差异,它们在不同物种中的保守作用。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Gene
生物-遗传学
CiteScore
6.10
自引率
2.90%
发文量
718
审稿时长
42 days
期刊介绍:
Gene publishes papers that focus on the regulation, expression, function and evolution of genes in all biological contexts, including all prokaryotic and eukaryotic organisms, as well as viruses.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


