Structure Prediction of Large RNAs with AlphaFold3 Highlights its Capabilities and Limitations
IF 4.7
2区 生物学
Q1 BIOCHEMISTRY & MOLECULAR BIOLOGY
引用次数: 0
Abstract
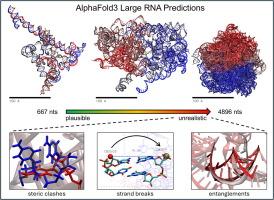
DeepMind’s AlphaFold3 webserver offers exciting new opportunities to make structural predictions of heterogeneous macromolecular systems. Here we attempt to apply AlphaFold3 to large RNA molecules whose 3D atomic structures are unknown but whose physical dimensions have been studied experimentally. One difficulty that we encounter is that models returned by AlphaFold3 often contain severe steric clashes and, less frequently, clear breaks in the phosphodiester backbone, with the probability of both events increasing with the length of the RNA. Restricting attention to those RNAs for which non-clashing models can be obtained, we find that hydrodynamic radii computed from the AlphaFold3 models are much larger than those reported experimentally under low salt conditions but are in better agreement with those reported in the presence of polyvalent cations. For two RNAs whose shapes have been imaged experimentally, the computed anisotropies of the AlphaFold3-predicted structures are too low, indicating that they are excessively spherical; extending this analysis to larger RNAs shows that they become progressively more spherical with increasing length. Overall, the results suggest that AlphaFold3 is capable of producing plausible models for RNAs up to ∼2000 nucleotides in length, but that thousands of predictions may be required to obtain models free of geometric problems.
利用 AlphaFold3 预测大型 RNA 的结构凸显了其能力和局限性。
DeepMind 的 AlphaFold3 网络服务器为异质大分子系统的结构预测提供了令人兴奋的新机会。在此,我们尝试将 AlphaFold3 应用于大型 RNA 分子,这些分子的三维原子结构尚不清楚,但其物理尺寸已经过实验研究。我们遇到的一个困难是,AlphaFold3 返回的模型经常包含严重的立体冲突,而且较少出现磷酸二酯骨架的明显断裂,这两种情况的发生概率随着 RNA 长度的增加而增加。我们将注意力限制在那些可以获得非冲突模型的 RNA 上,发现根据 AlphaFold3 模型计算出的流体力学半径远远大于低盐条件下的实验结果,但与多价阳离子存在时的实验结果更为一致。对于两种形状已通过实验成像的 RNA,AlphaFold3 预测结构的计算各向异性太低,表明它们过于球形;将这一分析扩展到更大的 RNA 表明,随着长度的增加,它们会逐渐变得更球形。总之,研究结果表明,AlphaFold3 能够为长度在 2000 个核苷酸以下的 RNA 生成可信的模型,但要获得不存在几何问题的模型,可能需要数千次预测。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Journal of Molecular Biology
生物-生化与分子生物学
CiteScore
11.30
自引率
1.80%
发文量
412
审稿时长
28 days
期刊介绍:
Journal of Molecular Biology (JMB) provides high quality, comprehensive and broad coverage in all areas of molecular biology. The journal publishes original scientific research papers that provide mechanistic and functional insights and report a significant advance to the field. The journal encourages the submission of multidisciplinary studies that use complementary experimental and computational approaches to address challenging biological questions.
Research areas include but are not limited to: Biomolecular interactions, signaling networks, systems biology; Cell cycle, cell growth, cell differentiation; Cell death, autophagy; Cell signaling and regulation; Chemical biology; Computational biology, in combination with experimental studies; DNA replication, repair, and recombination; Development, regenerative biology, mechanistic and functional studies of stem cells; Epigenetics, chromatin structure and function; Gene expression; Membrane processes, cell surface proteins and cell-cell interactions; Methodological advances, both experimental and theoretical, including databases; Microbiology, virology, and interactions with the host or environment; Microbiota mechanistic and functional studies; Nuclear organization; Post-translational modifications, proteomics; Processing and function of biologically important macromolecules and complexes; Molecular basis of disease; RNA processing, structure and functions of non-coding RNAs, transcription; Sorting, spatiotemporal organization, trafficking; Structural biology; Synthetic biology; Translation, protein folding, chaperones, protein degradation and quality control.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


