A multi-modal deep language model for contaminant removal from metagenome-assembled genomes
IF 18.8
1区 计算机科学
Q1 COMPUTER SCIENCE, ARTIFICIAL INTELLIGENCE
引用次数: 0
Abstract
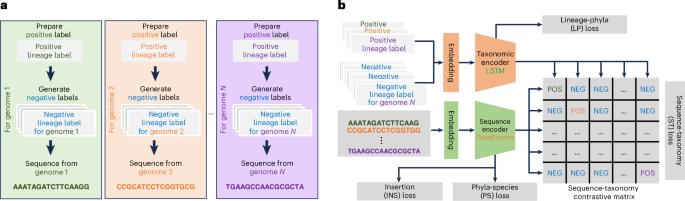
Metagenome-assembled genomes (MAGs) offer valuable insights into the exploration of microbial dark matter using metagenomic sequencing data. However, there is growing concern that contamination in MAGs may substantially affect the results of downstream analysis. Current MAG decontamination tools primarily rely on marker genes and do not fully use the contextual information of genomic sequences. To overcome this limitation, we introduce Deepurify for MAG decontamination. Deepurify uses a multi-modal deep language model with contrastive learning to match microbial genomic sequences with their taxonomic lineages. It allocates contigs within a MAG to a MAG-separated tree and applies a tree traversal algorithm to partition MAGs into sub-MAGs, with the goal of maximizing the number of high- and medium-quality sub-MAGs. Here we show that Deepurify outperformed MDMclearer and MAGpurify on simulated data, CAMI datasets and real-world datasets with varying complexities. Deepurify increased the number of high-quality MAGs by 20.0% in soil, 45.1% in ocean, 45.5% in plants, 33.8% in freshwater and 28.5% in human faecal metagenomic sequencing datasets. Metagenome-assembled genomes (MAGs) provide insights into microbial dark matter, but contamination remains a concern for downstream analysis. Zou et al. develop a multi-modal deep language model that leverages microbial sequences to remove ‘unexpected’ contigs from MAGs. This approach is compatible with any contig binning tools and increases the number of high-quality bins.
从元基因组组装基因组中清除污染物的多模式深度语言模型
元基因组组装基因组(MAGs)为利用元基因组测序数据探索微生物暗物质提供了宝贵的见解。然而,人们越来越担心,MAGs 中的污染可能会严重影响下游分析的结果。目前的 MAG 净化工具主要依赖标记基因,不能充分利用基因组序列的上下文信息。为了克服这一局限,我们推出了用于 MAG 去污的 Deepurify。Deepurify 使用具有对比学习功能的多模态深度语言模型来匹配微生物基因组序列及其分类学系谱。它将 MAG 中的等位基因分配到一棵 MAG 分离树上,并应用树遍历算法将 MAG 划分为子 MAG,目的是最大限度地增加高质量和中等质量子 MAG 的数量。在这里,我们展示了 Deepurify 在模拟数据、CAMI 数据集和具有不同复杂性的真实世界数据集上的表现优于 MDMclearer 和 MAGpurify。在土壤、海洋、植物、淡水和人类粪便元基因组测序数据集中,Deepurify 使高质量 MAG 的数量分别增加了 20.0%、45.1%、45.5%、33.8% 和 28.5%。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Nature Machine Intelligence
Multiple-
CiteScore
36.90
自引率
2.10%
发文量
127
期刊介绍:
Nature Machine Intelligence is a distinguished publication that presents original research and reviews on various topics in machine learning, robotics, and AI. Our focus extends beyond these fields, exploring their profound impact on other scientific disciplines, as well as societal and industrial aspects. We recognize limitless possibilities wherein machine intelligence can augment human capabilities and knowledge in domains like scientific exploration, healthcare, medical diagnostics, and the creation of safe and sustainable cities, transportation, and agriculture. Simultaneously, we acknowledge the emergence of ethical, social, and legal concerns due to the rapid pace of advancements.
To foster interdisciplinary discussions on these far-reaching implications, Nature Machine Intelligence serves as a platform for dialogue facilitated through Comments, News Features, News & Views articles, and Correspondence. Our goal is to encourage a comprehensive examination of these subjects.
Similar to all Nature-branded journals, Nature Machine Intelligence operates under the guidance of a team of skilled editors. We adhere to a fair and rigorous peer-review process, ensuring high standards of copy-editing and production, swift publication, and editorial independence.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


