Structural insights into the mechanism underlying the dual cofactor specificity of glyoxylate reductase from Acetobacter aceti in the β-hydroxyacid dehydrogenase family
IF 2.3
4区 生物学
Q3 BIOCHEMISTRY & MOLECULAR BIOLOGY
Biochimica et biophysica acta. Proteins and proteomics
Pub Date : 2024-10-03
DOI:10.1016/j.bbapap.2024.141051
引用次数: 0
Abstract
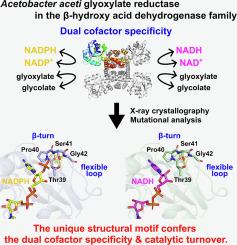
The β-hydroxyacid dehydrogenase family exhibits diverse cofactor preferences: some enzymes favor NAD, others favor NADP, and a subset can utilize both NAD and NADPH. Glyoxylate reductase from Acetobacter aceti JCM 20276 (AacGR) exhibits a dual cofactor specificity for NADPH and NADH in its catalytic reduction of glyoxylate to glycolate. In contrast to conventional cofactor-discriminating motifs, NRX and DXX, found in NADP- and NAD-specific enzymes, respectively, AacGR has a TPS motif in the equivalent position. Here we report X-ray crystallographic analysis of AacGR in its ligand-free form, and in complexes with NADPH and NADH, revealing critical interactions: Ser41 of the TPS motif interacted with the 2′-phosphate group of NADPH, while no analogous interaction occurred with the ribose hydroxy groups of NADH. Moreover, the TPS motif resided within a characteristic β-turn-like structure adjacent to a long flexible loop. Site-directed mutagenesis and kinetic analyses suggest that Ser41 facilitates NADPH binding, while the lack of a direct interaction of the TPS motif with NADH may allow for NADH utilization. The conformational dynamics of the TPS-containing β-turn-like structure along with the flexible loop likely govern the dual cofactor specificity and catalytic turnover of AacGR.
从结构上揭示β-羟基酸脱氢酶家族中乙酸醋酸杆菌乙醛酸还原酶的双辅助因子特异性机制。
β-羟基酸脱氢酶家族表现出多种多样的辅助因子偏好:一些酶偏好 NAD,另一些则偏好 NADP,还有一部分既能利用 NAD 也能利用 NADPH。醋酸纤维菌 JCM 20276(AacGR)的乙醛酸还原酶在催化乙醛酸还原为乙醇酸的过程中,表现出对 NADPH 和 NADH 的双重辅助因子特异性。与分别存在于 NADP 和 NAD 特异性酶中的传统辅因子区分基团 NRX 和 DXX 不同,AacGR 在同等位置上具有一个 TPS 基团。在这里,我们报告了对无配体形式的 AacGR 以及 AacGR 与 NADPH 和 NADH 复合物的 X 射线晶体学分析,揭示了关键的相互作用:TPS 主题的 Ser41 与 NADPH 的 2'- 磷酸基团相互作用,而与 NADH 的核糖羟基则没有类似的相互作用。此外,TPS基序位于一个特征性的β-turn-like结构中,毗邻一个长的柔性环。定点突变和动力学分析表明,Ser41 有助于 NADPH 的结合,而 TPS 基序与 NADH 之间缺乏直接相互作用,这可能会导致 NADH 的利用。含 TPS 的 β 转环结构和柔性环的构象动力学可能决定了 AacGR 的双辅助因子特异性和催化周转。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊
CiteScore
8.00
自引率
0.00%
发文量
55
审稿时长
33 days
期刊介绍:
BBA Proteins and Proteomics covers protein structure conformation and dynamics; protein folding; protein-ligand interactions; enzyme mechanisms, models and kinetics; protein physical properties and spectroscopy; and proteomics and bioinformatics analyses of protein structure, protein function, or protein regulation.

 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


