Monitoring clonal burden as an alternative to blast count for myelodysplastic neoplasm treatment response
IF 12.8
1区 医学
Q1 HEMATOLOGY
引用次数: 0
Abstract
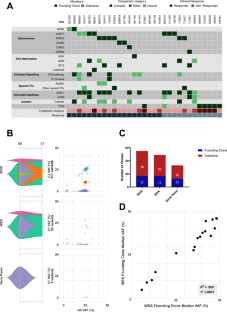
Accurate assessment of therapy response in myelodysplastic neoplasm (MDS) has been challenging. Directly monitoring mutational disease burden may be useful, but is not currently included in MDS response criteria, and the correlation of mutational burden and traditional response endpoints is not completely understood. Here, we used genome-wide and targeted next-generation sequencing (NGS) to monitor clonal and subclonal molecular disease burden in 452 samples from 32 patients prospectively treated in a clinical trial. Molecular responses were compared with International Working Group (IWG) 2006-defined response assessments. We found that myeloblast percentage consistently underestimates MDS molecular disease burden and that mutational clearance patterns for marrow complete remission (mCR), which depends on myeloblast assessment, was not different than stable disease or bone marrow aplasia, underscoring a major limitation of using mCR. In contrast, achieving a complete remission (CR) was associated with the highest level of mutation clearance and lowest residual mutational burden in higher-risk MDS patients. A targeted gene panel approach was inferior to genome-wide sequencing in defining subclones and their molecular responses but may be adequate for monitoring molecular disease burden when a targeted gene is present in the founding clone. Our work supports incorporating serial NGS-based monitoring into prospective MDS clinical trials.

监测克隆负担,替代囊泡计数,以了解骨髓增生异常肿瘤的治疗反应
准确评估骨髓增生异常肿瘤(MDS)的治疗反应一直是一项挑战。直接监测突变疾病负担可能有用,但目前尚未纳入 MDS 反应标准,而且突变负担与传统反应终点的相关性尚未完全明了。在这里,我们使用全基因组和靶向下一代测序(NGS)来监测临床试验中接受前瞻性治疗的 32 例患者的 452 份样本中的克隆和亚克隆分子疾病负担。分子反应与国际工作组(IWG)2006 年定义的反应评估进行了比较。我们发现,骨髓细胞百分比始终低估了 MDS 分子疾病负担,而骨髓完全缓解(mCR)的突变清除模式(取决于骨髓细胞评估)与疾病稳定或骨髓增生无异,这突出了使用 mCR 的一个主要局限性。相比之下,在高风险MDS患者中,获得完全缓解(CR)与最高的突变清除率和最低的残留突变负荷有关。在定义亚克隆及其分子反应方面,靶向基因面板方法不如全基因组测序方法,但当创始克隆中存在靶向基因时,靶向基因面板方法可能足以监测分子疾病负担。我们的工作支持将基于 NGS 的序列监测纳入前瞻性 MDS 临床试验。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Leukemia
医学-血液学
CiteScore
18.10
自引率
3.50%
发文量
270
审稿时长
3-6 weeks
期刊介绍:
Title: Leukemia
Journal Overview:
Publishes high-quality, peer-reviewed research
Covers all aspects of research and treatment of leukemia and allied diseases
Includes studies of normal hemopoiesis due to comparative relevance
Topics of Interest:
Oncogenes
Growth factors
Stem cells
Leukemia genomics
Cell cycle
Signal transduction
Molecular targets for therapy
And more
Content Types:
Original research articles
Reviews
Letters
Correspondence
Comments elaborating on significant advances and covering topical issues
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


