The structural landscape of Microprocessor-mediated processing of pri-let-7 miRNAs
IF 14.5
1区 生物学
Q1 BIOCHEMISTRY & MOLECULAR BIOLOGY
引用次数: 0
Abstract
MicroRNA (miRNA) biogenesis is initiated upon cleavage of a primary miRNA (pri-miRNA) hairpin by the Microprocessor (MP), composed of the Drosha RNase III enzyme and its partner DGCR8. Multiple pri-miRNA sequence motifs affect MP recognition, fidelity, and efficiency. Here, we performed cryoelectron microscopy (cryo-EM) and biochemical studies of several let-7 family pri-miRNAs in complex with human MP. We show that MP has the structural plasticity to accommodate a range of pri-miRNAs. These structures revealed key features of the 5′ UG sequence motif, more comprehensively represented as the “flipped U with paired N” (fUN) motif. Our analysis explains how cleavage of class-II pri-let-7 members harboring a bulged nucleotide generates a non-canonical precursor with a 1-nt 3′ overhang. Finally, the MP-SRSF3-pri-let-7f1 structure reveals how SRSF3 contributes to MP fidelity by interacting with the CNNC motif and Drosha’s Piwi/Argonaute/Zwille (PAZ)-like domain. Overall, this study sheds light on the mechanisms for flexible recognition, accurate cleavage, and regulated processing of different pri-miRNAs by MP.
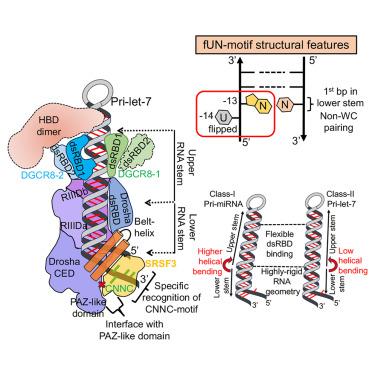
微处理器介导的 pri-let-7 miRNA 处理结构图
由 Drosha RNase III 酶及其伙伴 DGCR8 组成的微处理器(MP)对初级 miRNA(pri-miRNA)发夹进行切割后,就开始了微 RNA(miRNA)的生物生成。多种 pri-miRNA 序列基序会影响 MP 的识别、保真度和效率。在这里,我们对几种与人类 MP 复合的 let-7 家族 pri-miRNA 进行了冷冻电子显微镜(cryo-EM)和生化研究。我们发现 MP 具有结构可塑性,可以容纳一系列 pri-miRNA。这些结构揭示了 5′ UG 序列基序的关键特征,更全面地表述为 "带成对 N 的翻转 U"(fUN)基序。我们的分析解释了携带凸起核苷酸的 II 类 pri-let-7 成员如何通过裂解产生具有 1-nt 3′ 悬垂的非经典前体。最后,MP-SRSF3-pri-let-7f1结构揭示了SRSF3如何通过与CNNC基序和Drosha的Piwi/Argonaute/Zwille(PAZ)样结构域相互作用来促进MP的保真度。总之,这项研究揭示了 MP 灵活识别、准确切割和调控处理不同 pri-miRNA 的机制。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Molecular Cell
生物-生化与分子生物学
CiteScore
26.00
自引率
3.80%
发文量
389
审稿时长
1 months
期刊介绍:
Molecular Cell is a companion to Cell, the leading journal of biology and the highest-impact journal in the world. Launched in December 1997 and published monthly. Molecular Cell is dedicated to publishing cutting-edge research in molecular biology, focusing on fundamental cellular processes. The journal encompasses a wide range of topics, including DNA replication, recombination, and repair; Chromatin biology and genome organization; Transcription; RNA processing and decay; Non-coding RNA function; Translation; Protein folding, modification, and quality control; Signal transduction pathways; Cell cycle and checkpoints; Cell death; Autophagy; Metabolism.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


