FuseLinker: Leveraging LLM’s pre-trained text embeddings and domain knowledge to enhance GNN-based link prediction on biomedical knowledge graphs
IF 4
2区 医学
Q2 COMPUTER SCIENCE, INTERDISCIPLINARY APPLICATIONS
引用次数: 0
Abstract
Objective
To develop the FuseLinker, a novel link prediction framework for biomedical knowledge graphs (BKGs), which fully exploits the graph’s structural, textual and domain knowledge information. We evaluated the utility of FuseLinker in the graph-based drug repurposing task through detailed case studies.
Methods
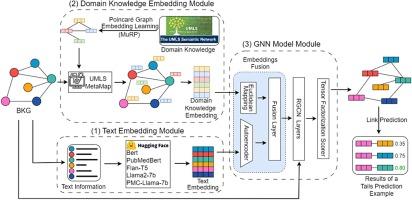
FuseLinker leverages fused pre-trained text embedding and domain knowledge embedding to enhance the graph neural network (GNN)-based link prediction model tailored for BKGs. This framework includes three parts: a) obtain text embeddings for BKGs using embedding-visible large language models (LLMs), b) learn the representations of medical ontology as domain knowledge information by employing the Poincaré graph embedding method, and c) fuse these embeddings and further learn the graph structure representations of BKGs by applying a GNN-based link prediction model. We evaluated FuseLinker against traditional knowledge graph embedding models and a conventional GNN-based link prediction model across four public BKG datasets. Additionally, we examined the impact of using different embedding-visible LLMs on FuseLinker’s performance. Finally, we investigated FuseLinker’s ability to generate medical hypotheses through two drug repurposing case studies for Sorafenib and Parkinson’s disease.
Results
By comparing FuseLinker with baseline models on four BKGs, our method demonstrates superior performance. The Mean Reciprocal Rank (MRR) and Area Under receiver operating characteristic Curve (AUROC) for KEGG50k, Hetionet, SuppKG and ADInt are 0.969 and 0.987, 0.548 and 0.903, 0.739 and 0.928, and 0.831 and 0.890, respectively.
Conclusion
Our study demonstrates that FuseLinker is an effective novel link prediction framework that integrates multiple graph information and shows significant potential for practical applications in biomedical and clinical tasks. Source code and data are available at https://github.com/YKXia0/FuseLinker.
FuseLinker:利用 LLM 的预训练文本嵌入和领域知识,增强基于 GNN 的生物医学知识图谱链接预测。
目的开发用于生物医学知识图谱(BKG)的新型链接预测框架FuseLinker,该框架可充分利用图谱的结构、文本和领域知识信息。我们通过详细的案例研究评估了 FuseLinker 在基于图的药物再利用任务中的实用性:FuseLinker 利用融合预训练文本嵌入和领域知识嵌入来增强基于图神经网络 (GNN) 的链接预测模型,该模型专为 BKGs 量身定制。该框架包括三个部分:a)使用嵌入可见大语言模型(LLM)获取 BKG 的文本嵌入;b)使用 Poincaré 图嵌入方法学习作为领域知识信息的医学本体表示;c)融合这些嵌入,并通过应用基于 GNN 的链接预测模型进一步学习 BKG 的图结构表示。我们通过四个公开的 BKG 数据集对 FuseLinker 与传统的知识图嵌入模型和传统的基于 GNN 的链接预测模型进行了评估。此外,我们还考察了使用不同的嵌入可见 LLM 对 FuseLinker 性能的影响。最后,我们通过索拉非尼(Sorafenib)和帕金森病的两个药物再利用案例研究,考察了 FuseLinker 生成医学假设的能力:结果:通过比较 FuseLinker 与四个 BKG 的基线模型,我们的方法表现出了卓越的性能。KEGG50k、Hetionet、SuppKG 和 ADInt 的平均倒数秩(MRR)和接收者操作特征曲线下面积(AUROC)分别为 0.969 和 0.987、0.548 和 0.903、0.739 和 0.928 以及 0.831 和 0.890:我们的研究表明,FuseLinker 是一种有效的新型链接预测框架,它整合了多种图信息,在生物医学和临床任务的实际应用中显示出巨大的潜力。源代码和数据见 https://github.com/YKXia0/FuseLinker。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Journal of Biomedical Informatics
医学-计算机:跨学科应用
CiteScore
8.90
自引率
6.70%
发文量
243
审稿时长
32 days
期刊介绍:
The Journal of Biomedical Informatics reflects a commitment to high-quality original research papers, reviews, and commentaries in the area of biomedical informatics methodology. Although we publish articles motivated by applications in the biomedical sciences (for example, clinical medicine, health care, population health, and translational bioinformatics), the journal emphasizes reports of new methodologies and techniques that have general applicability and that form the basis for the evolving science of biomedical informatics. Articles on medical devices; evaluations of implemented systems (including clinical trials of information technologies); or papers that provide insight into a biological process, a specific disease, or treatment options would generally be more suitable for publication in other venues. Papers on applications of signal processing and image analysis are often more suitable for biomedical engineering journals or other informatics journals, although we do publish papers that emphasize the information management and knowledge representation/modeling issues that arise in the storage and use of biological signals and images. System descriptions are welcome if they illustrate and substantiate the underlying methodology that is the principal focus of the report and an effort is made to address the generalizability and/or range of application of that methodology. Note also that, given the international nature of JBI, papers that deal with specific languages other than English, or with country-specific health systems or approaches, are acceptable for JBI only if they offer generalizable lessons that are relevant to the broad JBI readership, regardless of their country, language, culture, or health system.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


