BCL-Former: Localized Transformer Fusion with Balanced Constraint for polyp image segmentation
IF 7
2区 医学
Q1 BIOLOGY
引用次数: 0
Abstract
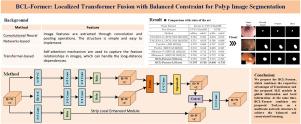
Polyp segmentation remains challenging for two reasons: (a) the size and shape of colon polyps are variable and diverse; (b) the distinction between polyps and mucosa is not obvious. To solve the above two challenging problems and enhance the generalization ability of segmentation method, we propose the Localized Transformer Fusion with Balanced Constraint (BCL-Former) for Polyp Segmentation. In BCL-Former, the Strip Local Enhancement module (SLE module) is proposed to capture the enhanced local features. The Progressive Feature Fusion module (PFF module) is presented to make the feature aggregation smoother and eliminate the difference between high-level and low-level features. Moreover, the Tversky-based Appropriate Constrained Loss (TacLoss) is proposed to achieve the balance and constraint between True Positives and False Negatives, improving the ability to generalize across datasets. Extensive experiments are conducted on four benchmark datasets. Results show that our proposed method achieves state-of-the-art performance in both segmentation precision and generalization ability. Also, the proposed method is 5%–8% faster than the benchmark method in training and inference. The code is available at: https://github.com/sjc-lbj/BCL-Former.
BCL-Former:用于息肉图像分割的具有平衡约束条件的局部变换器融合。
由于以下两个原因,息肉分割仍然具有挑战性:(a) 结肠息肉的大小和形状多变且多样化;(b) 息肉和粘膜之间的区别不明显。为了解决上述两个难题并提高分割方法的泛化能力,我们提出了用于息肉分割的均衡约束局部变换器融合(BCL-Former)方法。在 BCL-Former 中,我们提出了带状局部增强模块(SLE 模块)来捕捉增强的局部特征。此外,还提出了渐进式特征融合模块(PFF 模块),使特征聚合更加平滑,消除了高级特征和低级特征之间的差异。此外,还提出了基于 Tversky 的适当约束损失(TacLoss),以实现真阳性和假阴性之间的平衡和约束,提高跨数据集的泛化能力。我们在四个基准数据集上进行了广泛的实验。结果表明,我们提出的方法在分割精度和泛化能力方面都达到了最先进的水平。此外,所提出的方法在训练和推理方面比基准方法快 5%-8%。代码见:https://github.com/sjc-lbj/BCL-Former。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Computers in biology and medicine
工程技术-工程:生物医学
CiteScore
11.70
自引率
10.40%
发文量
1086
审稿时长
74 days
期刊介绍:
Computers in Biology and Medicine is an international forum for sharing groundbreaking advancements in the use of computers in bioscience and medicine. This journal serves as a medium for communicating essential research, instruction, ideas, and information regarding the rapidly evolving field of computer applications in these domains. By encouraging the exchange of knowledge, we aim to facilitate progress and innovation in the utilization of computers in biology and medicine.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


