Thermostable Nucleoid Protein Cren7 Slides Along DNA and Rapidly Dissociates From DNA While Not Inhibiting the Sliding of Other DNA-binding Protein
IF 4.7
2区 生物学
Q1 BIOCHEMISTRY & MOLECULAR BIOLOGY
引用次数: 0
Abstract
A nucleoid protein Cren7 compacts DNA, contributing to the living of Crenarchaeum in high temperature environment. In this study, we investigated the dynamic behavior of Cren7 on DNA and its functional relation using single-molecule fluorescence microscopy. We found two mobility modes of Cren7, sliding along DNA and pausing on it, and the rapid dissociation kinetics from DNA. The salt dependence analysis suggests a sliding with continuous contact to DNA, rather than hopping/jumping. The mutational analysis demonstrates that Cren7 slides along DNA while Trp (W26) residue interacts with the DNA. Furthermore, Cren7 does not impede the target search by a model transcription factor p53, implying no significant interference to other DNA-binding proteins on DNA. At high concentration of Cren7, the molecules form large clusters on DNA via bridging, which compacts DNA. We discuss how the dynamic behavior of Cren7 on DNA enables DNA-compaction and protein-bypass functions.
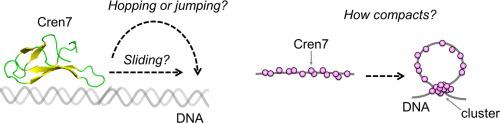
耐热核糖体蛋白 Cren7 可沿 DNA 滑动,并迅速与 DNA 分离,同时不会抑制其他 DNA 结合蛋白的滑动。
一种核壳蛋白 Cren7 可压实 DNA,从而促进了 Crenarchaeum 在高温环境中的生存。本研究利用单分子荧光显微镜研究了 Cren7 在 DNA 上的动态行为及其功能关系。我们发现了 Cren7 在 DNA 上的两种移动模式:沿 DNA 滑动和在 DNA 上停顿,以及与 DNA 快速解离的动力学。盐依赖性分析表明,Cren7 是一种与 DNA 持续接触的滑动模式,而不是跳跃模式。突变分析表明,当 Trp(W26)残基与 DNA 相互作用时,Cren7 沿着 DNA 滑动。此外,Cren7 并不妨碍模型转录因子 p53 的目标搜索,这意味着对 DNA 上的其他 DNA 结合蛋白没有明显干扰。在高浓度 Cren7 的情况下,分子通过桥接作用在 DNA 上形成大的簇,从而压实 DNA。我们讨论了 Cren7 在 DNA 上的动态行为是如何实现 DNA 压实和蛋白质旁路功能的。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Journal of Molecular Biology
生物-生化与分子生物学
CiteScore
11.30
自引率
1.80%
发文量
412
审稿时长
28 days
期刊介绍:
Journal of Molecular Biology (JMB) provides high quality, comprehensive and broad coverage in all areas of molecular biology. The journal publishes original scientific research papers that provide mechanistic and functional insights and report a significant advance to the field. The journal encourages the submission of multidisciplinary studies that use complementary experimental and computational approaches to address challenging biological questions.
Research areas include but are not limited to: Biomolecular interactions, signaling networks, systems biology; Cell cycle, cell growth, cell differentiation; Cell death, autophagy; Cell signaling and regulation; Chemical biology; Computational biology, in combination with experimental studies; DNA replication, repair, and recombination; Development, regenerative biology, mechanistic and functional studies of stem cells; Epigenetics, chromatin structure and function; Gene expression; Membrane processes, cell surface proteins and cell-cell interactions; Methodological advances, both experimental and theoretical, including databases; Microbiology, virology, and interactions with the host or environment; Microbiota mechanistic and functional studies; Nuclear organization; Post-translational modifications, proteomics; Processing and function of biologically important macromolecules and complexes; Molecular basis of disease; RNA processing, structure and functions of non-coding RNAs, transcription; Sorting, spatiotemporal organization, trafficking; Structural biology; Synthetic biology; Translation, protein folding, chaperones, protein degradation and quality control.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


