Cryo-EM Reveals the Mechanism of DNA Compaction by Mycobacterium smegmatis Dps2
IF 4.7
2区 生物学
Q1 BIOCHEMISTRY & MOLECULAR BIOLOGY
引用次数: 0
Abstract
DNA binding protein from starved cells (Dps) is a miniature ferritin complex, which plays a vital role in protecting bacterial DNA during starvation to maintain the integrity of bacteria under hostile conditions. Several approaches, including cryo-electron tomography, have been previously implemented by other research groups to decipher the structure of the Dps protein bound to DNA. However, none of the structures of the Dps-DNA complex was resolved to high resolution to identify the DNA binding residues. Like other bacteria, Mycobacterium smegmatis also expresses Dps2 (called MsDps2), which binds DNA to protect it under oxidative stress conditions. In this study, we implemented various biochemical and biophysical studies to characterize the DNA protein interactions of Dps2 protein from Mycobacterium smegmatis. We employed single-particle cryo-EM-based structural analysis of MsDps2-DNA complexes and identified that the region close to the N-terminus confers the DNA binding property. Based on cryo-EM data, we also pinpointed several arginine residues, proximal to the DNA binding region, responsible for DNA binding. We also performed mutations of these residues, which dramatically reduced the MsDps2-DNA interaction. In addition, we proposed a model that elucidates the mechanism of DNA compaction, which adapts a lattice-like structure. We performed single-molecule imaging of MsDps2-DNA interactions that corroborate well with our structural studies. Taken together, our results delineate the specific MsDps2 residues that play an important role in DNA binding and compaction, providing new insights into Mycobacterial DNA compaction mechanisms under stress conditions.
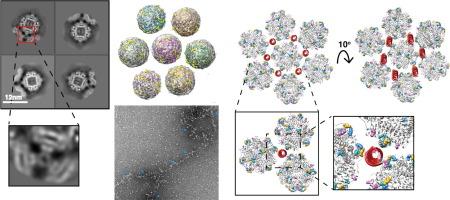
低温电子显微镜揭示了分枝杆菌 Dps2 的 DNA 压实机制。
饥饿细胞的 DNA 结合蛋白(Dps)是一种微型铁蛋白复合物,在饥饿过程中保护细菌 DNA 方面发挥着重要作用,从而在恶劣条件下保持细菌的完整性。此前,其他研究小组采用了包括低温电子断层扫描在内的多种方法来破译 Dps 蛋白与 DNA 结合的结构。然而,没有一个 Dps-DNA 复合物的结构被解析到高分辨率,无法确定 DNA 结合残基。与其他细菌一样,分枝杆菌也表达 Dps2(称为 MsDps2),它与 DNA 结合,在氧化压力条件下保护 DNA。在本研究中,我们进行了各种生化和生物物理研究,以确定烟曲霉分枝杆菌 Dps2 蛋白的 DNA 蛋白相互作用的特征。我们采用单颗粒低温电子显微镜对 MsDps2-DNA 复合物进行了结构分析,发现靠近 N 端的区域具有 DNA 结合特性。根据低温电子显微镜数据,我们还确定了几个精氨酸残基,它们位于 DNA 结合区的近端,负责 DNA 结合。我们还对这些残基进行了突变,结果大大降低了 MsDps2 与 DNA 的相互作用。此外,我们还提出了一个模型,该模型阐明了 DNA 的压实机制,它适应了一种类似晶格的结构。我们对 MsDps2-DNA 的相互作用进行了单分子成像,结果与我们的结构研究完全吻合。综上所述,我们的研究结果确定了在 DNA 结合和压实过程中发挥重要作用的特定 MsDps2 残基,为我们深入了解分枝杆菌 DNA 在应激条件下的压实机制提供了新的视角。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Journal of Molecular Biology
生物-生化与分子生物学
CiteScore
11.30
自引率
1.80%
发文量
412
审稿时长
28 days
期刊介绍:
Journal of Molecular Biology (JMB) provides high quality, comprehensive and broad coverage in all areas of molecular biology. The journal publishes original scientific research papers that provide mechanistic and functional insights and report a significant advance to the field. The journal encourages the submission of multidisciplinary studies that use complementary experimental and computational approaches to address challenging biological questions.
Research areas include but are not limited to: Biomolecular interactions, signaling networks, systems biology; Cell cycle, cell growth, cell differentiation; Cell death, autophagy; Cell signaling and regulation; Chemical biology; Computational biology, in combination with experimental studies; DNA replication, repair, and recombination; Development, regenerative biology, mechanistic and functional studies of stem cells; Epigenetics, chromatin structure and function; Gene expression; Membrane processes, cell surface proteins and cell-cell interactions; Methodological advances, both experimental and theoretical, including databases; Microbiology, virology, and interactions with the host or environment; Microbiota mechanistic and functional studies; Nuclear organization; Post-translational modifications, proteomics; Processing and function of biologically important macromolecules and complexes; Molecular basis of disease; RNA processing, structure and functions of non-coding RNAs, transcription; Sorting, spatiotemporal organization, trafficking; Structural biology; Synthetic biology; Translation, protein folding, chaperones, protein degradation and quality control.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


