Toward subtask-decomposition-based learning and benchmarking for predicting genetic perturbation outcomes and beyond
IF 12
Q1 COMPUTER SCIENCE, INTERDISCIPLINARY APPLICATIONS
引用次数: 0
Abstract
Deciphering cellular responses to genetic perturbations is fundamental for a wide array of biomedical applications. However, there are three main challenges: predicting single-genetic-perturbation outcomes, predicting multiple-genetic-perturbation outcomes and predicting genetic outcomes across cell lines. Here we introduce Subtask Decomposition Modeling for Genetic Perturbation Prediction (STAMP), a flexible artificial intelligence strategy for genetic perturbation outcome prediction and downstream applications. STAMP formulates genetic perturbation prediction as a subtask decomposition problem by resolving three progressive subtasks in a problem decomposition manner, that is, identifying postperturbation differentially expressed genes, determining the expression change directions of differentially expressed genes and finally estimating the magnitudes of gene expression changes. STAMP exhibits a substantial improvement over the existing approaches on three subtasks and beyond, including the ability to identify key regulatory genes and pathways on small samples and to reveal precise genetic interactions of diverse types. By employing the subtask decomposition strategy, STAMP outperforms existing models in single, multiple and cross-cell-line scenarios for genetic perturbation prediction, showing potential to uncover gene regulations and interactions.
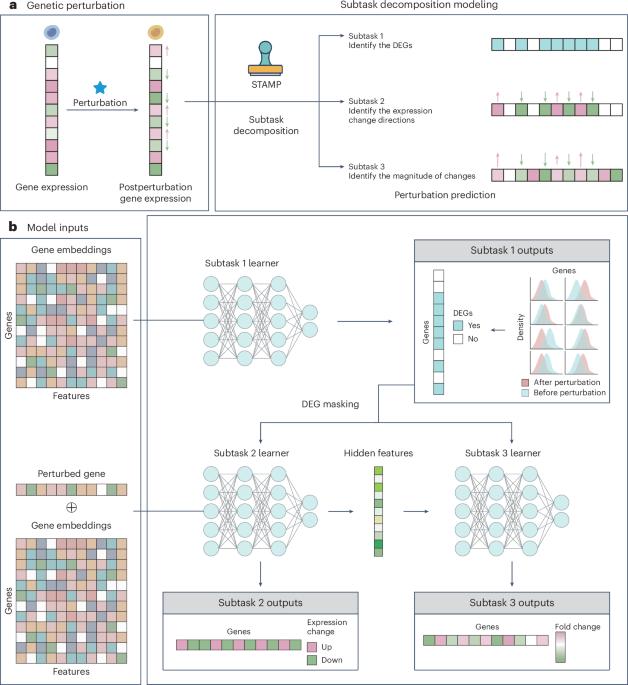
基于子任务分解的学习和基准,用于预测遗传扰动结果及其他。
破译细胞对遗传扰动的反应是广泛生物医学应用的基础。然而,目前存在三大挑战:预测单基因扰动结果、预测多基因扰动结果和预测跨细胞系基因结果。在此,我们介绍用于遗传扰动预测的子任务分解模型(STAMP),这是一种灵活的人工智能策略,可用于遗传扰动结果预测和下游应用。STAMP 将遗传扰动预测表述为一个子任务分解问题,以问题分解的方式解决三个渐进的子任务,即识别扰动后差异表达基因、确定差异表达基因的表达变化方向以及最后估计基因表达变化的幅度。与现有方法相比,STAMP 在三个子任务及其他方面都有很大改进,包括能够识别小样本中的关键调控基因和通路,精确揭示不同类型的遗传相互作用。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文

 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


