Uncovering Intermolecular Interactions Driving the Liquid–Liquid Phase Separation of the TDP-43 Low-Complexity Domain via Atomistic Dimerization Simulations
IF 5.6
2区 化学
Q1 CHEMISTRY, MEDICINAL
引用次数: 0
Abstract
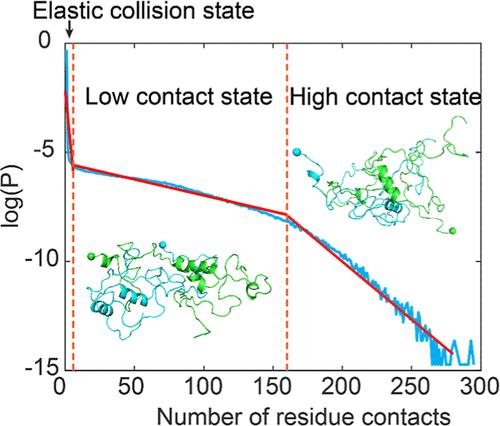
Liquid–liquid phase separation (LLPS) of transactive response DNA-binding protein of 43 kDa (TDP-43), which exerts multiple functions in the splicing, trafficking, and stabilization of RNA, mediates the formation of membraneless condensates with crucial physiological roles, while its aberrant LLPS is linked to multiple neurodegenerative diseases. However, due to the heterogeneous and dynamic nature of LLPS, major gaps remain in understanding the precise intermolecular interactions driving LLPS and how specific mutations alter LLPS dynamics. Here, we investigated the molecular mechanisms underlying the LLPS of the TDP-43 low-complexity domain (LCD) by simulating the dimerization process using all-atom discrete molecular dynamics with microsecond-long simulations. Our results showed that the TDP-43 LCD was intrinsically disordered, with helical structures consistent with prior nuclear magnetic resonance studies. Phase separation propensity was assessed by simulating the dimerization of the TDP-43 LCD and four mutants, showing that A321G, W334G, and M337V inhibited self-association, while G335D promoted it, fully consistent with experimental reports. During the dimerization process, two peptides experienced both elastic and nonelastic collisions, and the self-associated dimer featured both high- and low-contact states. These results suggested that the dimerization process of the TDP-43 LCD was accordingly dynamic and heterogeneous. Additionally, we identified crucial regions containing hydrophobic clusters and aromatic residues in the N-terminus, central region, and C-terminus that were essential for the self-association of the TDP-43 LCD. These residues with high binding affinities can act as stickers to form peptide networks in LLPS. Together, our simulation provides a comprehensive picture of the intermolecular interactions driving the phase separation of the TDP-43 LCD, offering insights into both physiological functions and pathological mechanisms.
通过原子二聚化模拟揭示驱动 TDP-43 低复杂性结构域液-液相分离的分子间相互作用
转录反应 DNA 结合蛋白 43 kDa(TDP-43)的液-液相分离(LLPS)在 RNA 的剪接、贩运和稳定过程中发挥多种功能,它介导无膜凝聚物的形成,具有重要的生理作用,而其异常的 LLPS 与多种神经退行性疾病有关。然而,由于 LLPS 的异质性和动态性,在理解驱动 LLPS 的精确分子间相互作用以及特定突变如何改变 LLPS 动态方面仍存在重大差距。在这里,我们通过使用全原子离散分子动力学微秒长模拟二聚化过程,研究了 TDP-43 低复杂度结构域(LCD)LLPS 的分子机制。我们的结果表明,TDP-43 的 LCD 本质上是无序的,其螺旋结构与之前的核磁共振研究一致。通过模拟 TDP-43 LCD 和四种突变体的二聚化,评估了相分离倾向,结果显示 A321G、W334G 和 M337V 抑制了自结合,而 G335D 则促进了自结合,这与实验报告完全一致。在二聚过程中,两条肽经历了弹性和非弹性碰撞,自结合的二聚体具有高接触和低接触两种状态。这些结果表明,TDP-43 LCD 的二聚过程具有相应的动态性和异质性。此外,我们还在 TDP-43 LCD 的 N 端、中心区和 C 端发现了包含疏水簇和芳香残基的关键区域,这些残基对 TDP-43 LCD 的自结合至关重要。这些具有高结合亲和力的残基可以作为粘合剂,在 LLPS 中形成肽网络。总之,我们的模拟全面描述了驱动 TDP-43 LCD 相分离的分子间相互作用,为生理功能和病理机制提供了见解。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊
CiteScore
9.80
自引率
10.70%
发文量
529
审稿时长
1.4 months
期刊介绍:
The Journal of Chemical Information and Modeling publishes papers reporting new methodology and/or important applications in the fields of chemical informatics and molecular modeling. Specific topics include the representation and computer-based searching of chemical databases, molecular modeling, computer-aided molecular design of new materials, catalysts, or ligands, development of new computational methods or efficient algorithms for chemical software, and biopharmaceutical chemistry including analyses of biological activity and other issues related to drug discovery.
Astute chemists, computer scientists, and information specialists look to this monthly’s insightful research studies, programming innovations, and software reviews to keep current with advances in this integral, multidisciplinary field.
As a subscriber you’ll stay abreast of database search systems, use of graph theory in chemical problems, substructure search systems, pattern recognition and clustering, analysis of chemical and physical data, molecular modeling, graphics and natural language interfaces, bibliometric and citation analysis, and synthesis design and reactions databases.

 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


