Talking about diseases; developing a model of patient and public-prioritised disease phenotypes
IF 12.4
1区 医学
Q1 HEALTH CARE SCIENCES & SERVICES
引用次数: 0
Abstract
Deep phenotyping describes the use of standardised terminologies to create comprehensive phenotypic descriptions of biomedical phenomena. These characterisations facilitate secondary analysis, evidence synthesis, and practitioner awareness, thereby guiding patient care. The vast majority of this knowledge is derived from sources that describe an academic understanding of disease, including academic literature and experimental databases. Previous work indicates a gulf between the priorities, perspectives, and perceptions held by different healthcare stakeholders. Using social media data, we develop a phenotype model that represents a public perspective on disease and compare this with a model derived from a combination of existing academic phenotype databases. We identified 52,198 positive disease-phenotype associations from social media across 311 diseases. We further identified 24,618 novel phenotype associations not shared by the biomedical and literature-derived phenotype model across 304 diseases, of which we considered 14,531 significant. Manifestations of disease affecting quality of life, and concerning endocrine, digestive, and reproductive diseases were over-represented in the social media phenotype model. An expert clinical review found that social media-derived associations were considered similarly well-established to those derived from literature, and were seen significantly more in patient clinical encounters. The phenotype model recovered from social media presents a significantly different perspective than existing resources derived from biomedical databases and literature, providing a large number of associations novel to the latter dataset. We propose that the integration and interrogation of these public perspectives on the disease can inform clinical awareness, improve secondary analysis, and bridge understanding and priorities across healthcare stakeholders.
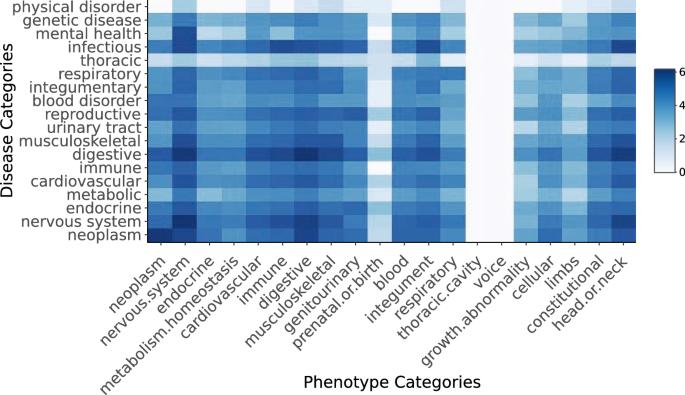
谈论疾病;建立患者和公众优先考虑的疾病表型模型
深度表型描述的是使用标准化术语对生物医学现象进行全面的表型描述。这些特征描述有助于二次分析、证据综合和从业人员认知,从而指导患者护理。这些知识绝大多数来自对疾病的学术理解,包括学术文献和实验数据库。以往的研究表明,不同的医疗保健利益相关者所持有的优先级、观点和看法之间存在差距。利用社交媒体数据,我们建立了一个代表公众对疾病看法的表型模型,并将其与从现有学术表型数据库组合中得出的模型进行比较。我们从社交媒体的 311 种疾病中发现了 52198 种疾病与表型之间的正相关。我们还在 304 种疾病中发现了 24,618 种生物医学和文献表型模型所不具备的新表型关联,我们认为其中 14,531 种具有重要意义。在社交媒体表型模型中,影响生活质量的疾病表现,以及与内分泌、消化和生殖疾病有关的疾病表现所占比例较高。专家临床审查发现,从社交媒体中得出的关联与从文献中得出的关联具有相似性,而且在患者临床会诊中出现的关联明显更多。与生物医学数据库和文献中的现有资源相比,从社交媒体中恢复的表型模型呈现出明显不同的视角,为后者的数据集提供了大量新颖的关联。我们建议,对这些公众对疾病的看法进行整合和分析,可以为临床认识提供信息,改善二次分析,并在医疗保健利益相关者之间架起理解和优先事项的桥梁。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

NPJ Digital Medicine
Multiple-
CiteScore
25.10
自引率
3.30%
发文量
170
审稿时长
15 weeks
期刊介绍:
npj Digital Medicine is an online open-access journal that focuses on publishing peer-reviewed research in the field of digital medicine. The journal covers various aspects of digital medicine, including the application and implementation of digital and mobile technologies in clinical settings, virtual healthcare, and the use of artificial intelligence and informatics.
The primary goal of the journal is to support innovation and the advancement of healthcare through the integration of new digital and mobile technologies. When determining if a manuscript is suitable for publication, the journal considers four important criteria: novelty, clinical relevance, scientific rigor, and digital innovation.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


