Predicting Collision-Induced-Dissociation Tandem Mass Spectra (CID-MS/MS) Using Ab Initio Molecular Dynamics
IF 5.3
2区 化学
Q1 CHEMISTRY, MEDICINAL
引用次数: 0
Abstract
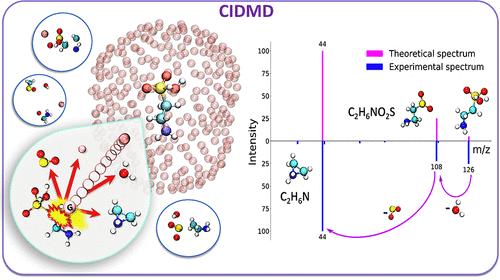
Compound identification is at the center of metabolomics, usually by comparing experimental mass spectra against library spectra. However, most compounds are not commercially available to generate library spectra. Hence, for such compounds, MS/MS spectra need to be predicted. Machine learning and heuristic models have largely failed except for lipids. Here, quantum chemistry software can be used to predict mass spectra. However, quantum chemistry predictions for collision induced dissociation (CID) mass spectra in LC-MS/MS are rare. We present the CIDMD (Collision-Induced Dissociation via Molecular Dynamics) framework to model CID-based MS/MS spectra. It uses first-principles molecular dynamics (MD) to simulate the physical process of molecular collisions in CID tandem mass spectrometry. First, molecular ions are constructed at specific protonation sites. Using density functional theory, these protonated ions are targeted by argon collider gas atoms at user-specified velocities. Subsequent bond breakages are simulated over time for at least 1,000 fs. Each simulation is repeated multiple times from various collisional directions. Fragmentations are accumulated over those repeated collisions to generate CIDMD in silico mass spectra. Twelve small metabolites (<205 Da) were selected to test the accuracy of this framework in comparison to experimental MS/MS spectra. When testing different protomers, collider velocities, number of simulations, simulation time and impact factor b cutoffs, we yielded 261 predicted mass spectra. These in silico spectra resulted in entropy similarity scores of an average 624 ± 189 for all 261 spectra compared to their corresponding experimental spectra, which improved to 828 ± 77 when using optimal parameters of the most probable protomers for 12 molecules. With increasing molecular mass, higher velocities achieved better results. Similarly, different protomers showed large differences in fragmentation; hence, with increasing numbers of protomers and tautomers, the average CIDMD prediction accuracy decreased. Mechanistic details showed that specific fragment ions can be produced from different protomers via multiple fragmentation pathways. We propose that CIDMD is a suitable tool to predict mass spectra of small metabolites like produced by the gut microbiome.
利用 Ab Initio 分子动力学预测碰撞诱导解离串联质谱 (CID-MS/MS)
化合物鉴定是代谢组学的核心,通常是将实验质谱与库谱进行比较。然而,大多数化合物都无法通过商业途径生成库谱。因此,需要对这些化合物的 MS/MS 图谱进行预测。除了脂质之外,机器学习和启发式模型基本上都失败了。在这方面,量子化学软件可用于预测质谱。然而,LC-MS/MS 中碰撞诱导解离(CID)质谱的量子化学预测却很少见。我们提出了 CIDMD(Collision-Induced Dissociation via Molecular Dynamics,通过分子动力学的碰撞诱导解离)框架来模拟基于 CID 的 MS/MS 图谱。它采用第一原理分子动力学(MD)模拟 CID 串联质谱中分子碰撞的物理过程。首先,在特定质子化位点构建分子离子。利用密度泛函理论,这些质子化离子以用户指定的速度成为氩气对撞机气体原子的目标。随后的键断裂模拟时间至少为 1,000 fs。每次模拟都会从不同的碰撞方向重复多次。碎片在这些重复碰撞中累积,生成 CIDMD 硅质谱。我们选择了 12 种小型代谢物(205 Da)来测试该框架与实验 MS/MS 质谱比较的准确性。在测试不同的原生质体、对撞机速度、模拟次数、模拟时间和影响因子 b 截止值时,我们得到了 261 个预测质谱。与相应的实验光谱相比,所有 261 个光谱的熵相似性得分平均为 624 ± 189,当使用 12 个分子最可能的原质体的最佳参数时,相似性得分提高到 828 ± 77。随着分子质量的增加,速度越高,结果越好。同样,不同的原生体在片段化方面也表现出很大差异;因此,随着原生体和同系物数量的增加,CIDMD 的平均预测准确度也在下降。机理细节表明,不同的原生质体可以通过多种碎片途径产生特定的碎片离子。我们认为 CIDMD 是预测肠道微生物组产生的小型代谢物质谱的合适工具。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊
CiteScore
9.80
自引率
10.70%
发文量
529
审稿时长
1.4 months
期刊介绍:
The Journal of Chemical Information and Modeling publishes papers reporting new methodology and/or important applications in the fields of chemical informatics and molecular modeling. Specific topics include the representation and computer-based searching of chemical databases, molecular modeling, computer-aided molecular design of new materials, catalysts, or ligands, development of new computational methods or efficient algorithms for chemical software, and biopharmaceutical chemistry including analyses of biological activity and other issues related to drug discovery.
Astute chemists, computer scientists, and information specialists look to this monthly’s insightful research studies, programming innovations, and software reviews to keep current with advances in this integral, multidisciplinary field.
As a subscriber you’ll stay abreast of database search systems, use of graph theory in chemical problems, substructure search systems, pattern recognition and clustering, analysis of chemical and physical data, molecular modeling, graphics and natural language interfaces, bibliometric and citation analysis, and synthesis design and reactions databases.

 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


