Enhancing solution structural analysis of large molecular proteins through optimal stereo array isotope labeling of aromatic amino acids
IF 3.3
3区 生物学
Q2 BIOCHEMISTRY & MOLECULAR BIOLOGY
引用次数: 0
Abstract
The observation of side-chain peaks of aromatic amino acids is the prerequisite for a high-resolution three-dimensional structure determination of proteins by NMR. However, it becomes difficult with increasing molecular size due to an increased transverse relaxation and the control of the relaxation pathway is needed to achieve the observation. We demonstrated that even for the large molecular size of 82 kDa Malate synthase G (MSG), the aromatic 13C-1H (CH) peaks of Tryptophan (Trp) and Phenylalanine (Phe) residues can be observed with high quality using a systematic stable isotope labeling scheme, Stereo-Array Isotope Labeling (SAIL) method. However, the sequence specific assignments of these peaks relied on the use of amino acid substitutions, employing an inefficient method that required many isotopes labeled samples. In this study, we developed novel SAIL amino acids that allow for the observation of the aromatic ring δ,ζ and the aliphatic β position peak of Phe residues. The application of TROSY-based experiment to the isolated CH moieties resulted in the successful observation of discernible and resolved CH peaks in Phe residues in MSG. In MSG, the sequence-specific assignments of the backbone and Cβ positions have already been confirmed. Therefore, using this labeling method, the δ and β position peaks of Phe residues can be clearly assigned in a sequence-specific and stereospecific manner through experiments based on intra-residue NOE. Furthermore, the NOESY experiment also allows for the acquisition of information pertaining to the conformation of Phe residues, such as the χ1 dihedral angle, providing valuable insights for the determination of accurate protein structures and in dynamic analysis. This new SAIL amino acids open an avenue to achieve a variety of NMR analysis of large molecular proteins, including a high-resolution structure determination and dynamics and interaction analysis.
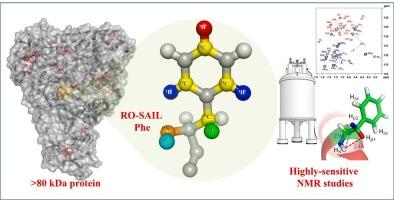
通过优化芳香族氨基酸的立体阵列同位素标记,加强大分子蛋白质的溶液结构分析
观察芳香族氨基酸的侧链峰是通过核磁共振确定蛋白质高分辨率三维结构的先决条件。然而,由于横向弛豫的增加,随着分子尺寸的增大,观测变得越来越困难,因此需要控制弛豫途径来实现观测。我们利用系统的稳定同位素标记方案--立体阵列同位素标记(SAIL)方法,证明了即使对于 82 kDa 的大分子苹果酸合成酶 G(MSG),也能高质量地观察到色氨酸(Trp)和苯丙氨酸(Phe)残基的芳香 13C-1H (CH) 峰。然而,这些峰的序列特异性分配依赖于氨基酸的替换,采用这种方法效率低下,需要大量同位素标记样品。在本研究中,我们开发了新型 SAIL 氨基酸,可以观察 Phe 残基的芳香环 δ、ζ 和脂肪族 β 位置峰。将基于 TROSY 的实验应用于分离的 CH 分子,成功地在味精中的 Phe 残基中观察到了可分辨和解析的 CH 峰。在味精中,骨架和 Cβ 位置的序列特异性分配已经得到证实。因此,利用这种标记方法,可以通过基于残基内 NOE 的实验,以序列特异性和立体特异性的方式明确分配 Phe 残基的 δ 和 β 位置峰。此外,NOESY 实验还能获取与 Phe 残基构象有关的信息,如 χ1 二面角,为确定准确的蛋白质结构和进行动态分析提供有价值的见解。这种新的 SAIL 氨基酸为实现大分子蛋白质的各种核磁共振分析(包括高分辨率结构测定和动力学及相互作用分析)开辟了一条途径。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊
Biophysical chemistry
生物-生化与分子生物学
CiteScore
6.10
自引率
10.50%
发文量
121
审稿时长
20 days
期刊介绍:
Biophysical Chemistry publishes original work and reviews in the areas of chemistry and physics directly impacting biological phenomena. Quantitative analysis of the properties of biological macromolecules, biologically active molecules, macromolecular assemblies and cell components in terms of kinetics, thermodynamics, spatio-temporal organization, NMR and X-ray structural biology, as well as single-molecule detection represent a major focus of the journal. Theoretical and computational treatments of biomacromolecular systems, macromolecular interactions, regulatory control and systems biology are also of interest to the journal.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


