Comparative analyses of the transcriptome among three development stages of Zeugodacus tau larvae (Diptera: Tephritidae)
IF 2.2
2区 生物学
Q4 BIOCHEMISTRY & MOLECULAR BIOLOGY
Comparative Biochemistry and Physiology D-Genomics & Proteomics
Pub Date : 2024-09-23
DOI:10.1016/j.cbd.2024.101333
引用次数: 0
Abstract
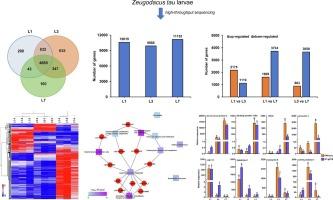
Studying differences in transcriptomes across various development stages of insects is necessary to uncover the physiological and molecular mechanism underlying development and metamorphosis. We here present the first transcriptome data generated under Illumina Hiseq platform concerning Zeugodacus tau (Walker) larvae from Nanchang, China. In total, 11,702 genes were identified from 9 transcriptome libraries of three development stages of Z. tau larvae. 7219 differentially expressed genes (DEGs) were screened out from the comparisons between each two development stages of Z. tau larvae, and their roles in development and metabolism were analyzed. Comparative analyses of transcriptome data showed that there are 5333 DEGs between 1-day and 7-day old larvae, consisting of 1609 up-regulated and 3724 down-regulated genes. Expressions of DEGs were more abundant in L7 than in L1 and L3, which might be associated with metamorphosis. Gene Ontology (GO) enrichment and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis suggested the enrichment of metabolic process. KOG annotation further confirmed that 20-hydroxyecdysone (20E) pathway related genes Cyp4ac1_1, Cyp4aa1, Cyp313a4_3 were critical for the biosynthesis, transport, and catabolism of secondary metabolites and lipid transport and metabolism. Expression patterns of 8 DEGs were verified using quantitative real-time PCR (RT-qPCR). This study elucidated the DEGs and their roles underlying three development stages of Z. tau larvae, which provided valuable information for further functional genomic research.
Zeugodacus tau 幼虫(双翅目:头螨科)三个发育阶段转录组的比较分析
研究昆虫不同发育阶段转录组的差异对于揭示昆虫发育和变态的生理和分子机制十分必要。在此,我们首次展示了在 Illumina Hiseq 平台下产生的有关中国南昌的 Zeugodacus tau (Walker) 幼虫的转录组数据。在三个发育阶段的 9 个转录组文库中,我们共鉴定出 11702 个基因。从每两个发育阶段的差异表达基因(DEGs)中筛选出 7219 个差异表达基因,并分析了它们在发育和代谢过程中的作用。转录组数据的比较分析表明,1日龄和7日龄幼虫共有5333个DEGs,其中上调基因1609个,下调基因3724个。与 L1 和 L3 相比,DEGs 在 L7 的表达更为丰富,这可能与变态有关。基因本体(GO)富集和京都基因和基因组百科全书(KEGG)通路分析表明,代谢过程富集。KOG 注释进一步证实,20-羟基蜕皮激素(20E)通路相关基因 Cyp4ac1_1、Cyp4aa1、Cyp313a4_3 对次级代谢产物的生物合成、运输和分解以及脂质运输和代谢至关重要。利用实时定量 PCR(RT-qPCR)验证了 8 个 DEGs 的表达模式。该研究阐明了舟尾幼虫三个发育阶段的DEGs及其作用,为进一步的功能基因组研究提供了宝贵的信息。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊
CiteScore
5.10
自引率
3.30%
发文量
69
审稿时长
33 days
期刊介绍:
Comparative Biochemistry & Physiology (CBP) publishes papers in comparative, environmental and evolutionary physiology.
Part D: Genomics and Proteomics (CBPD), focuses on “omics” approaches to physiology, including comparative and functional genomics, metagenomics, transcriptomics, proteomics, metabolomics, and lipidomics. Most studies employ “omics” and/or system biology to test specific hypotheses about molecular and biochemical mechanisms underlying physiological responses to the environment. We encourage papers that address fundamental questions in comparative physiology and biochemistry rather than studies with a focus that is purely technical, methodological or descriptive in nature.

 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


