Taxonomic identification of Morocco scorpions using MALDI-MS fingerprints of venom proteomes and computational modeling
IF 2.8
2区 生物学
Q2 BIOCHEMICAL RESEARCH METHODS
引用次数: 0
Abstract
The venom of scorpions has been the subject of numerous studies. However, their taxonomic identification is not a simple task, leading to misidentifications. This study aims to provide a practical approach for identifying scorpions based on the venom molecular mass fingerprint (MFP). Specimens (251) belonging to fifteen species were collected from different regions in Morocco. Their MFPs were acquired using MALDI-MS. These were used as a training dataset to generate predictive models and a library of mean spectral profiles using software programs based on machine learning. The computational model achieved an overall recognition capability of 99 % comprising 32 molecular signatures. The models and the library were tested using a new dataset for external validation and to evaluate their capability of identification. We recorded an accuracy classification with an average of 97 % and 98 % for the computational models and the library, respectively. To our knowledge, this is the first attempt to demonstrate the potential of MALDI-MS and MFPs to generate predictive models capable of discriminating scorpions from family to species levels, and to build a library of species-specific spectra. These promising results may represent a proof of concept towards developing a reliable approach for rapid molecular identification of scorpions in Morocco.
Significance of the study
With their clinical importance, scorpions may constitute a desirable study model for many researchers. The first step in studying scorpion is systematically identifying the species of interest. However, it can be a difficult task, especially for the non-experts. The taxonomy of scorpions is primarily based on morphometric characters. In Morocco, the high number of species and subspecies mainly endemic, and the morphological similarities between different species may result in false identifications. This was observed in many reports according to the scorpion experts. In this study, we describe a reliable practical approach for identifying scorpions based on the venom molecular mass fingerprints (MFPs). By using two software programs based on machine learning, we have demonstrated that these MFPs contains sufficient inter-specific variation to differentiate between the scorpion species mentioned in this study with a good accuracy. Using a drop of venom, this new approach could be a rapid, accurate and cost saving method for taxonomic identification of scorpions in Morocco.
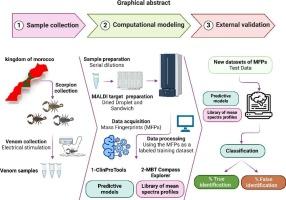
利用毒液蛋白质组的 MALDI-MS 指纹和计算模型对摩洛哥蝎子进行分类鉴定。
蝎子的毒液一直是众多研究的主题。然而,蝎子毒液的分类鉴定并不简单,容易造成误认。本研究旨在提供一种基于毒液分子质量指纹(MFP)鉴定蝎子的实用方法。研究人员从摩洛哥不同地区采集了属于 15 个物种的标本(251 个)。使用 MALDI-MS 获取了它们的分子质量指纹。这些标本被用作训练数据集,利用基于机器学习的软件程序生成预测模型和平均谱图库。计算模型的总体识别能力达到 99%,包括 32 个分子特征。我们使用一个新的数据集对模型和资料库进行了测试,以进行外部验证并评估其识别能力。我们发现,计算模型和库的平均分类准确率分别为 97% 和 98%。据我们所知,这是首次尝试证明 MALDI-MS 和 MFPs 在生成预测模型方面的潜力,这些模型能够对蝎子从科到种进行鉴别,并建立物种特异性光谱库。这些充满希望的结果可能代表了一种概念验证,即开发一种可靠的方法来快速分子鉴定摩洛哥的蝎子。研究意义:由于蝎子在临床上的重要性,它们可能成为许多研究人员理想的研究模型。研究蝎子的第一步是系统地鉴定感兴趣的物种。然而,这可能是一项艰巨的任务,尤其是对非专业人员而言。蝎子的分类主要基于形态特征。在摩洛哥,主要是特有种和亚种的数量很多,不同物种之间的形态相似性可能会导致错误的识别。蝎子专家在许多报告中都发现了这种情况。在本研究中,我们介绍了一种基于毒液分子质量指纹(MFPs)的可靠实用的蝎子鉴定方法。通过使用两个基于机器学习的软件程序,我们证明了这些分子质量指纹包含足够的种间差异,能够准确地区分本研究中提到的蝎子种类。利用一滴毒液,这种新方法可以快速、准确和节省成本地对摩洛哥的蝎子进行分类鉴定。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Journal of proteomics
生物-生化研究方法
CiteScore
7.10
自引率
3.00%
发文量
227
审稿时长
73 days
期刊介绍:
Journal of Proteomics is aimed at protein scientists and analytical chemists in the field of proteomics, biomarker discovery, protein analytics, plant proteomics, microbial and animal proteomics, human studies, tissue imaging by mass spectrometry, non-conventional and non-model organism proteomics, and protein bioinformatics. The journal welcomes papers in new and upcoming areas such as metabolomics, genomics, systems biology, toxicogenomics, pharmacoproteomics.
Journal of Proteomics unifies both fundamental scientists and clinicians, and includes translational research. Suggestions for reviews, webinars and thematic issues are welcome.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


