Ontogeny, not prey availability, underlies allopatric venom variability in insular and mainland populations of Vipera ammodytes
IF 2.8
2区 生物学
Q2 BIOCHEMICAL RESEARCH METHODS
引用次数: 0
Abstract
Allopatric populations living under distinct ecological conditions are excellent systems to infer factors underlying intraspecific venom variation. The venom composition of two populations of Vipera ammodytes, insular with a diet based on ectotherms and mainland with a diet based on ectotherms and endotherms, was compared considering the sex and age of individuals. Ten toxin families, dominated by PLA2, svMP, svSP, and DI, were identified through a bottom-up approach. The venom profiles of adult females and males were similar. Results from 58 individual SDS-PAGE profiles and venom pool analysis revealed significant differences between juveniles compared to subadults and adults. Two venom phenotypes were identified: a juvenile svMP-dominated and KUN-lacking phenotype and an adult PLA2/svMP-balanced and KUN-containing phenotype. Despite differences in prey availability (and, therefore, diet) between populations, no significant differences in venom composition were found. As the populations are geographically isolated, the lack of venom diversification could be explained by insufficient time for natural selection and/or genetic drift to act on the venom composition of island vipers. However, substantial differences in proteomes were observed when compared to venoms from geographically distant populations inhabiting different conditions. These findings highlight the need to consider ecological and evolutionary processes when studying venom variability.
Significance
This study provides the first comprehensive analysis of the venom composition of two allopatric populations of Vipera ammodytes, living under similar abiotic (climate) but distinct biotic (prey availability) conditions. The ontogenetic changes in venom composition, coupled with the lack of differences between sex and between populations, shed light on the main determinants of venom evolution in this medically important snake. Seven new proteomes may facilitate future comparative studies of snake venom evolution. This study highlights the importance of considering ecological and evolutionary factors to understand snake venom variation.
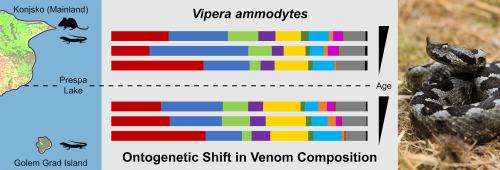
海岛和大陆蝰蛇种群异地毒液变异的基础是个体发育,而不是猎物的可获得性。
生活在不同生态条件下的同域种群是推断种内毒液变异因素的绝佳系统。考虑到个体的性别和年龄,研究人员比较了两个蝰蛇种群的毒液组成,一个是以外温动物为食的海岛种群,另一个是以外温动物和内温动物为食的大陆种群。通过自下而上的方法,确定了以 PLA2、svMP、svSP 和 DI 为主的 10 个毒素家族。成年雌性和雄性的毒液特征相似。58 个个体的 SDS-PAGE 图谱和毒液池分析结果显示,幼体与亚成体和成体相比存在显著差异。发现了两种毒液表型:一种是幼体以 svMP 为主、缺乏 KUN 的表型,另一种是成体 PLA2/svMP平衡、含有 KUN 的表型。尽管不同种群之间的猎物(因此也包括食物)存在差异,但毒液成分却没有发现明显的不同。由于这些种群在地理上是孤立的,毒液缺乏多样性的原因可能是自然选择和/或遗传漂移作用于岛屿毒蛇毒液组成的时间不足。然而,与来自地理位置遥远、生活环境不同的种群的毒液相比,蛋白质组存在很大差异。这些发现强调了在研究毒液变异性时考虑生态和进化过程的必要性。意义:本研究首次全面分析了生活在类似非生物(气候)但不同生物(猎物可用性)条件下的两个异地蝰蛇种群的毒液组成。毒液成分的个体发育变化以及不同性别和不同种群之间的差异,揭示了这种在医学上具有重要意义的蛇类毒液进化的主要决定因素。七个新的蛋白质组可能有助于未来对蛇毒进化的比较研究。这项研究强调了考虑生态和进化因素对理解蛇毒变异的重要性。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Journal of proteomics
生物-生化研究方法
CiteScore
7.10
自引率
3.00%
发文量
227
审稿时长
73 days
期刊介绍:
Journal of Proteomics is aimed at protein scientists and analytical chemists in the field of proteomics, biomarker discovery, protein analytics, plant proteomics, microbial and animal proteomics, human studies, tissue imaging by mass spectrometry, non-conventional and non-model organism proteomics, and protein bioinformatics. The journal welcomes papers in new and upcoming areas such as metabolomics, genomics, systems biology, toxicogenomics, pharmacoproteomics.
Journal of Proteomics unifies both fundamental scientists and clinicians, and includes translational research. Suggestions for reviews, webinars and thematic issues are welcome.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


