Comprehensive genetic analysis by targeted sequencing identifies risk factors and predicts patient outcome in Mantle Cell Lymphoma: results from the EU-MCL network trials
IF 12.8
1区 医学
Q1 HEMATOLOGY
引用次数: 0
Abstract
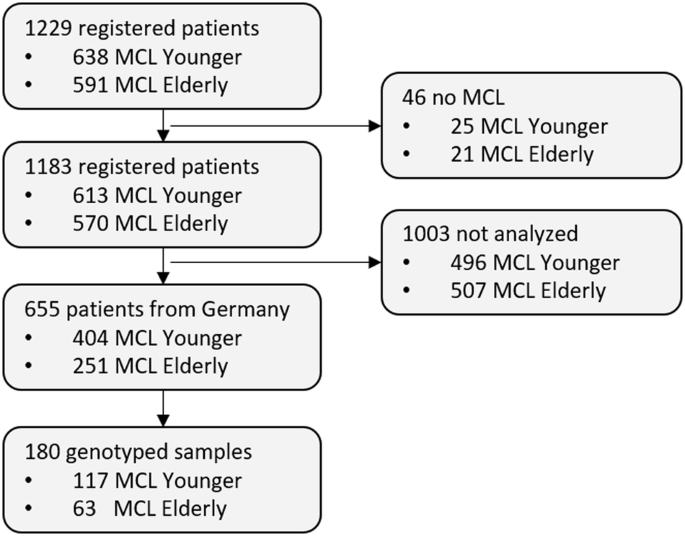
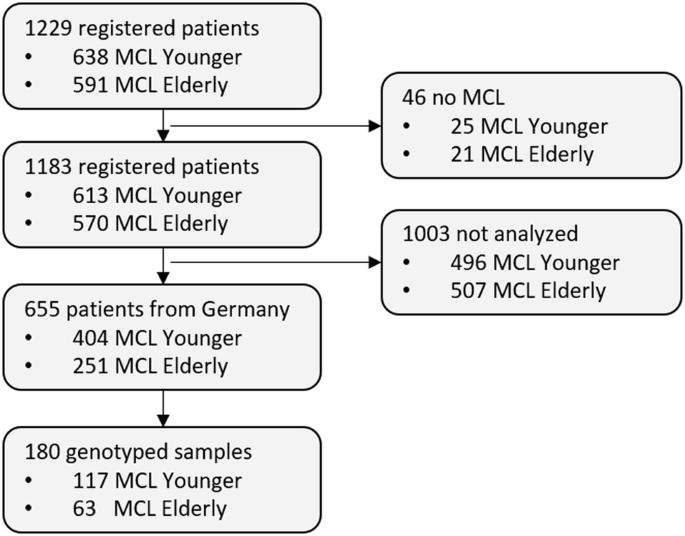
Recent studies highlighted genetic aberrations associated with prognosis in Mantle Cell lymphoma (MCL), yet comprehensive testing is not implemented in clinical routine. We conducted a comprehensive genomic characterization of 180 patients from the European MCL network trials by targeted sequencing of peripheral blood DNA using the EuroClonality(EC)-NDC assay. The IGH::CCND1 fusion was identified in 94% of patients, clonal IGH-V-(D)-J rearrangements in all, and 79% had ≥1 somatic gene mutation. The top mutated genes were ATM, TP53, KMT2D, SAMHD1, BIRC3 and NFKBIE. Copy number variations (CNVs) were detected in 83% of patients with RB1, ATM, CDKN2A/B and TP53 being the most frequently deleted and KLF2, CXCR4, CCND1, MAP2K1 and MYC the top amplified genes. CNVs and mutations were more frequently observed in older patients with adverse impact on prognosis. TP53mut, NOTCH1mut, FAT1mut TRAF2del, CDKN2A/Bdel and MAP2K1amp were linked to inferior failure-free (FFS) and overall survival (OS), while TRAF2mut, EGR2del and BCL2amp related to inferior OS only. Genetic complexity (≥3 CNVs) observed in 51% of analysed patients was significantly associated with impaired FFS and OS. We demonstrate that targeted sequencing from peripheral blood and bone marrow reliably detects diagnostically and prognostically important genetic factors in MCL patients, facilitating genetic characterization in clinical routine.
通过靶向测序进行综合基因分析,确定套细胞淋巴瘤的风险因素并预测患者预后:欧盟-套细胞淋巴瘤网络试验的结果
最近的研究强调了与套细胞淋巴瘤(MCL)预后相关的基因畸变,但全面的检测并未在临床常规中实施。我们使用EuroClonality(EC)-NDC测定法对外周血DNA进行了靶向测序,对欧洲MCL网络试验的180名患者进行了全面的基因组特征描述。94%的患者发现了IGH::CCND1融合,所有患者都发现了克隆性IGH-V-(D)-J重排,79%的患者有≥1个体细胞基因突变。突变基因最多的是ATM、TP53、KMT2D、SAMHD1、BIRC3和NFKBIE。83%的患者检测到拷贝数变异(CNV),其中RB1、ATM、CDKN2A/B和TP53是最常被删除的基因,KLF2、CXCR4、CCND1、MAP2K1和MYC是最常扩增的基因。CNV和基因突变在年龄较大的患者中更为常见,对预后有不利影响。TP53突变、NOTCH1突变、FAT1突变、TRAF2del、CDKN2A/Bdel和MAP2K1amp与较差的无失败(FFS)和总生存(OS)有关,而TRAF2突变、EGR2del和BCL2amp仅与较差的OS有关。在51%的分析患者中观察到的遗传复杂性(≥3个CNV)与无失败生存率和OS的降低有显著关系。我们证明,外周血和骨髓的靶向测序能可靠地检测出MCL患者在诊断和预后方面的重要遗传因素,从而促进临床常规的遗传特征描述。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Leukemia
医学-血液学
CiteScore
18.10
自引率
3.50%
发文量
270
审稿时长
3-6 weeks
期刊介绍:
Title: Leukemia
Journal Overview:
Publishes high-quality, peer-reviewed research
Covers all aspects of research and treatment of leukemia and allied diseases
Includes studies of normal hemopoiesis due to comparative relevance
Topics of Interest:
Oncogenes
Growth factors
Stem cells
Leukemia genomics
Cell cycle
Signal transduction
Molecular targets for therapy
And more
Content Types:
Original research articles
Reviews
Letters
Correspondence
Comments elaborating on significant advances and covering topical issues
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


