Exploring changes in social spider DNA methylation profiles in all cytosine contexts following infection
IF 3.9
2区 生物学
Q2 ECOLOGY
引用次数: 0
Abstract
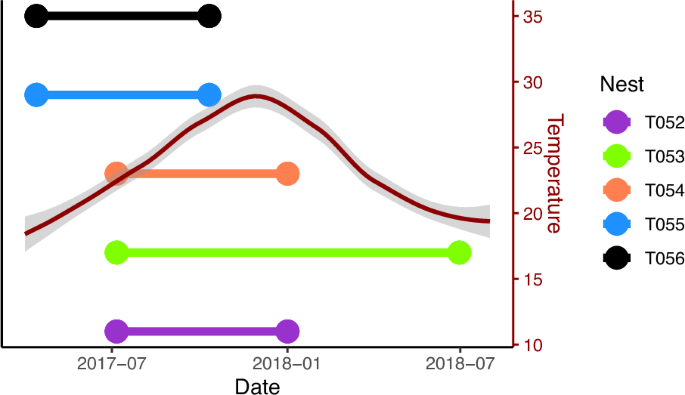
Living at high density and with low genetic diversity are factors that should both increase the susceptibility of organisms to disease. Therefore, group living organisms, especially those that are inbred, should be especially vulnerable to infection and therefore have particular strategies to cope with infection. Phenotypic plasticity, underpinned by epigenetic changes, could allow group living organisms to rapidly respond to infection challenges. To explore the potential role of epigenetic modifications in the immune response to a group-living species with low genetic diversity, we compared the genome-wide DNA methylation profiles of five colonies of social spiders (Stegodyphus dumicola) in their natural habitat in Namibia at the point just before they succumbed to infection to a point at least six months previously where they were presumably healthier. We found increases in genome- and chromosome-wide methylation levels in the CpG, CHG, and CHH contexts, although the genome-wide changes were not clearly different from zero. These changes were most prominent in the CHG context, especially at a narrow region of chromosome 13, hinting at an as-of-yet unsuspected role of this DNA methylation context in phenotypic plasticity. However, there were few clear patterns of differential methylation at the base level, and genes with a known immune function in spiders had mean methylation changes close to zero. Our results suggest that DNA methylation may change with infection at large genomic scales, but that this type of epigenetic change is not necessarily integral to the immune response of social spiders.
探索社会蜘蛛 DNA 甲基化图谱在感染后所有胞嘧啶上下文中的变化
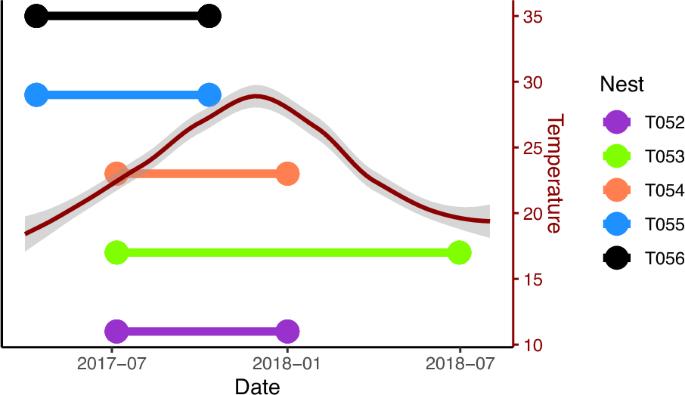
生活在高密度和低遗传多样性环境中的生物对疾病的易感性都会增加。因此,群居生物,尤其是那些近亲繁殖的生物,应该特别容易受到感染,并因此拥有应对感染的特殊策略。表型可塑性以表观遗传变化为基础,可使群居生物快速应对感染挑战。为了探索表观遗传修饰在对遗传多样性较低的群居物种的免疫反应中可能发挥的作用,我们比较了纳米比亚自然栖息地中五群社会蜘蛛(Stegodyphus dumicola)的全基因组 DNA 甲基化图谱。我们发现,在 CpG、CHG 和 CHH 背景下,基因组和整个染色体的甲基化水平都有所增加,尽管整个基因组的变化与零没有明显差异。这些变化在 CHG 上下文中最为突出,尤其是在 13 号染色体的一个狭窄区域,这暗示了这种 DNA 甲基化上下文在表型可塑性中尚未被发现的作用。然而,在碱基水平上几乎没有明显的甲基化差异模式,而且已知具有蜘蛛免疫功能的基因的平均甲基化变化接近于零。我们的研究结果表明,DNA甲基化可能会随着大基因组尺度的感染而发生变化,但这种类型的表观遗传变化并不一定与社会蜘蛛的免疫反应密不可分。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Heredity
生物-进化生物学
CiteScore
7.50
自引率
2.60%
发文量
84
审稿时长
4-8 weeks
期刊介绍:
Heredity is the official journal of the Genetics Society. It covers a broad range of topics within the field of genetics and therefore papers must address conceptual or applied issues of interest to the journal''s wide readership
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


