Multilevel Systematic Optimization To Achieve Efficient Integrated Expression of Escherichia coli
IF 3.7
2区 生物学
Q1 BIOCHEMICAL RESEARCH METHODS
引用次数: 0
Abstract
Genomic integration of heterologous genes is the preferred approach in industrial fermentation-related strains due to the drawbacks associated with plasmid-mediated microbial fermentation, including additional growth burden, genetic instability, and antibiotic contamination. Synthetic biology and genome editing advancements have made gene integration convenient. Integrated expression is extensively used in the field of biomanufacturing and is anticipated to become the prevailing method for expressing recombinant proteins. Therefore, it is pivotal to strengthen the expression of exogenous genes at the genome level. Here, we systematically optimized the integrated expression system of Escherichia coli from 3 aspects. First, the integration site slmA with the highest expression activity was screened out of 18 sites in the ORI region of the E. coli BL21 (DE3) genome. Second, we characterized 16 endogenous promoters in E. coli and combined them with the T7 promoter. A constitutive promoter, Plpp-T7, exhibited significantly higher expression strength than the T7 promoter, achieving a 3.3-fold increase in expression levels. Finally, to further enhance the T7 expression system, we proceeded with overexpression of T7 RNA polymerase at the chassis cell level. The resulting constitutive efficient integrated expression system (CEIES_Ecoli) showed a 2-fold increase in GFP expression compared to the pET3b recombinant plasmid. Therefore, CEIES_Ecoli was applied to the integrated expression of nitrilase and hyaluronidase, achieving stable and efficient enzyme expression, with enzyme activities of 22.87 and 12,195 U·mL–1, respectively, comparable to plasmid levels. Overall, CEIES_Ecoli provides a stable and efficient method of gene expression without the need for antibiotics or inducers, making it a robust tool for synthetic biology, enzyme engineering, and related applications.
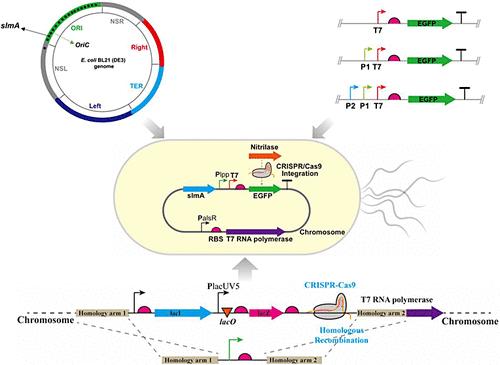
多级系统优化实现大肠杆菌的高效综合表达
异源基因的基因组整合是工业发酵相关菌株的首选方法,因为质粒介导的微生物发酵存在一些缺点,包括额外的生长负担、遗传不稳定性和抗生素污染。合成生物学和基因组编辑技术的进步为基因整合提供了便利。整合表达已广泛应用于生物制造领域,预计将成为表达重组蛋白的主流方法。因此,加强外源基因在基因组水平的表达至关重要。在此,我们从三个方面对大肠杆菌的整合表达系统进行了系统优化。首先,我们从大肠杆菌 BL21 (DE3) 基因组 ORI 区域的 18 个整合位点中筛选出了表达活性最高的整合位点 slmA。其次,我们鉴定了大肠杆菌中的 16 个内源启动子,并将它们与 T7 启动子相结合。组成型启动子 Plpp-T7 的表达强度明显高于 T7 启动子,表达水平提高了 3.3 倍。最后,为了进一步增强 T7 表达系统,我们继续在底盘细胞水平上过表达 T7 RNA 聚合酶。与 pET3b 重组质粒相比,由此产生的组成型高效整合表达系统(CEIES_Ecoli)的 GFP 表达量提高了 2 倍。因此,CEIES_Ecoli 被应用于硝化酶和透明质酸酶的整合表达,实现了稳定高效的酶表达,酶活分别为 22.87 和 12 195 U-mL-1,与质粒水平相当。总之,CEIES_Ecoli 提供了一种稳定高效的基因表达方法,无需抗生素或诱导剂,是合成生物学、酶工程和相关应用的有力工具。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊
CiteScore
8.00
自引率
10.60%
发文量
380
审稿时长
6-12 weeks
期刊介绍:
The journal is particularly interested in studies on the design and synthesis of new genetic circuits and gene products; computational methods in the design of systems; and integrative applied approaches to understanding disease and metabolism.
Topics may include, but are not limited to:
Design and optimization of genetic systems
Genetic circuit design and their principles for their organization into programs
Computational methods to aid the design of genetic systems
Experimental methods to quantify genetic parts, circuits, and metabolic fluxes
Genetic parts libraries: their creation, analysis, and ontological representation
Protein engineering including computational design
Metabolic engineering and cellular manufacturing, including biomass conversion
Natural product access, engineering, and production
Creative and innovative applications of cellular programming
Medical applications, tissue engineering, and the programming of therapeutic cells
Minimal cell design and construction
Genomics and genome replacement strategies
Viral engineering
Automated and robotic assembly platforms for synthetic biology
DNA synthesis methodologies
Metagenomics and synthetic metagenomic analysis
Bioinformatics applied to gene discovery, chemoinformatics, and pathway construction
Gene optimization
Methods for genome-scale measurements of transcription and metabolomics
Systems biology and methods to integrate multiple data sources
in vitro and cell-free synthetic biology and molecular programming
Nucleic acid engineering.
文献相关原料
| 公司名称 | 产品信息 | 采购帮参考价格 |
|---|

 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


