Probing the functional significance of wild animal microbiomes using omics data
IF 4.6
1区 环境科学与生态学
Q1 ECOLOGY
引用次数: 0
Abstract
Read the free Plain Language Summary for this article on the Journal blog.
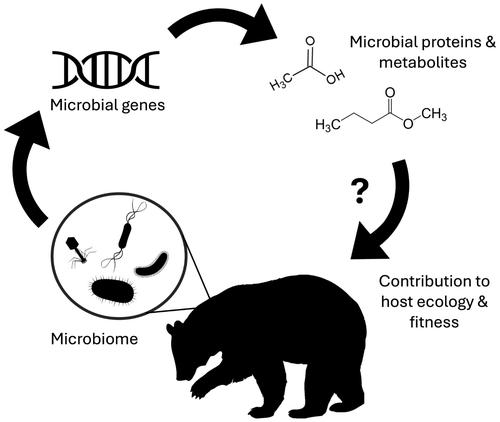
利用组学数据探究野生动物微生物组的功能意义
宿主相关微生物群被认为在宿主的生理和健康方面发挥着关键作用,但这一结论主要来自偏重于动物模型和人类的系统。虽然许多关于非模型动物和野生动物的研究都描述了其微生物组的分类多样性,但很少有研究对这些微生物群落的功能潜力进行调查。元基因组学、元转录组学和代谢组学等功能性 "omics "方法是探索野生宿主相关微生物组意义的有前途的技术。在本综述中,我们建议:(1) 简要定义现有的主要功能组学工具及其优势和局限性;(2) 总结组学工具在了解人类和动物模型微生物组功能方面取得的主要进展;(3) 举例说明这些方法如何为野生宿主微生物组带来了宝贵的见解;(4) 就如何利用这些工具解决野生动物微生物组领域的未决问题提供指导。最后,我们建议,在从更便宜、更传统的方法(如 16S 代谢编码和 qPCR)中获得的知识基础上,功能性 omics 工具是一种很有前途的方法,可用于检验有关野生动物常驻微生物群的生态和进化意义的假设。在期刊博客上免费阅读本文的通俗摘要。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Functional Ecology
环境科学-生态学
CiteScore
9.00
自引率
1.90%
发文量
243
审稿时长
4 months
期刊介绍:
Functional Ecology publishes high-impact papers that enable a mechanistic understanding of ecological pattern and process from the organismic to the ecosystem scale. Because of the multifaceted nature of this challenge, papers can be based on a wide range of approaches. Thus, manuscripts may vary from physiological, genetics, life-history, and behavioural perspectives for organismal studies to community and biogeochemical studies when the goal is to understand ecosystem and larger scale ecological phenomena. We believe that the diverse nature of our journal is a strength, not a weakness, and we are open-minded about the variety of data, research approaches and types of studies that we publish. Certain key areas will continue to be emphasized: studies that integrate genomics with ecology, studies that examine how key aspects of physiology (e.g., stress) impact the ecology of animals and plants, or vice versa, and how evolution shapes interactions among function and ecological traits. Ecology has increasingly moved towards the realization that organismal traits and activities are vital for understanding community dynamics and ecosystem processes, particularly in response to the rapid global changes occurring in earth’s environment, and Functional Ecology aims to publish such integrative papers.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


