{"title":"Genetic Manipulation of Candida glabrata","authors":"Jane Usher","doi":"10.1002/cpz1.70014","DOIUrl":null,"url":null,"abstract":"<p><i>Candida glabrata</i> (<i>Nakaseomyces glabratus</i>) is an opportunistic fungal pathogen that has become a significant concern in clinical settings due to its increasing resistance to antifungal treatments. Understanding the genetic basis of its pathogenicity and resistance mechanisms is crucial for developing new therapeutic strategies. One powerful method of studying gene function is through targeted gene deletion. This paper outlines a comprehensive protocol for the deletion of genes in <i>C. glabrata</i>, encompassing primer design, preparation of electrocompetent cells, transformation, and finally confirmation of the gene deletion. The protocol begins with the identification and design of primers necessary for generating deletion constructs, involving the precise targeting of up- and downstream regions flanking the gene of interest to ensure high specificity and efficiency of homologous recombination. Followed is the preparation of electrocompetent cells, a critical step for successful transformation. Transformation of the competent cells is achieved through electroporation, facilitating the introduction of exogenous DNA into the cells. This is followed by the selection and confirmation of successfully transformed colonies. Confirmation involves the use of colony PCR to verify the correct integration of the NAT resistance cassette and deletion of the target gene. © 2024 The Author(s). Current Protocols published by Wiley Periodicals LLC.</p><p><b>Basic Protocol 1</b>: Primer design for gene deletion in <i>C. glabrata</i></p><p><b>Basic Protocol 2</b>: Preparing competent <i>C. glabrata</i> cells</p><p><b>Basic Protocol 3</b>: Transforming <i>C. glabrata</i> using electroporation</p><p><b>Basic Protocol 4</b>: Confirming deletion strains with colony PCR</p>","PeriodicalId":93970,"journal":{"name":"Current protocols","volume":"4 9","pages":""},"PeriodicalIF":0.0000,"publicationDate":"2024-09-06","publicationTypes":"Journal Article","fieldsOfStudy":null,"isOpenAccess":false,"openAccessPdf":"https://onlinelibrary.wiley.com/doi/epdf/10.1002/cpz1.70014","citationCount":"0","resultStr":null,"platform":"Semanticscholar","paperid":null,"PeriodicalName":"Current protocols","FirstCategoryId":"1085","ListUrlMain":"https://onlinelibrary.wiley.com/doi/10.1002/cpz1.70014","RegionNum":0,"RegionCategory":null,"ArticlePicture":[],"TitleCN":null,"AbstractTextCN":null,"PMCID":null,"EPubDate":"","PubModel":"","JCR":"","JCRName":"","Score":null,"Total":0}
引用次数: 0
Abstract
Candida glabrata (Nakaseomyces glabratus) is an opportunistic fungal pathogen that has become a significant concern in clinical settings due to its increasing resistance to antifungal treatments. Understanding the genetic basis of its pathogenicity and resistance mechanisms is crucial for developing new therapeutic strategies. One powerful method of studying gene function is through targeted gene deletion. This paper outlines a comprehensive protocol for the deletion of genes in C. glabrata, encompassing primer design, preparation of electrocompetent cells, transformation, and finally confirmation of the gene deletion. The protocol begins with the identification and design of primers necessary for generating deletion constructs, involving the precise targeting of up- and downstream regions flanking the gene of interest to ensure high specificity and efficiency of homologous recombination. Followed is the preparation of electrocompetent cells, a critical step for successful transformation. Transformation of the competent cells is achieved through electroporation, facilitating the introduction of exogenous DNA into the cells. This is followed by the selection and confirmation of successfully transformed colonies. Confirmation involves the use of colony PCR to verify the correct integration of the NAT resistance cassette and deletion of the target gene. © 2024 The Author(s). Current Protocols published by Wiley Periodicals LLC.
Basic Protocol 1: Primer design for gene deletion in C. glabrata
Basic Protocol 2: Preparing competent C. glabrata cells
Basic Protocol 3: Transforming C. glabrata using electroporation
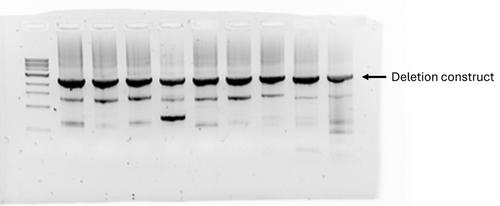
Basic Protocol 4: Confirming deletion strains with colony PCR
遗传操作光滑念珠菌。
胶状念珠菌(Nakaseomyces glabratus)是一种机会性真菌病原体,由于其对抗真菌治疗的耐药性不断增加,已成为临床环境中的一个重要问题。了解其致病性和抗药性机制的基因基础对于开发新的治疗策略至关重要。研究基因功能的一种有效方法是通过靶向基因缺失。本文概述了一种全面的草履虫基因缺失方案,包括引物设计、电能力细胞制备、转化以及最后的基因缺失确认。该方案首先要确定和设计生成基因缺失构建体所需的引物,包括精确瞄准感兴趣基因侧翼的上下游区域,以确保同源重组的高特异性和高效性。接下来是制备电能力细胞,这是成功转化的关键步骤。通过电穿孔实现合格细胞的转化,便于将外源 DNA 导入细胞。随后是筛选和确认成功转化的菌落。确认包括使用菌落 PCR 验证 NAT 抗性盒的正确整合和目标基因的缺失。© 2024 作者。当前协议》由 Wiley Periodicals LLC 出版。基本方案 1:设计引物以删除草履虫基因 基本方案 2:制备有能力的草履虫细胞 基本方案 3:使用电穿孔技术转化草履虫 基本方案 4:使用菌落 PCR 确认删除菌株。
本文章由计算机程序翻译,如有差异,请以英文原文为准。


 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


