Learning motif-based graphs for drug–drug interaction prediction via local–global self-attention
IF 18.8
1区 计算机科学
Q1 COMPUTER SCIENCE, ARTIFICIAL INTELLIGENCE
引用次数: 0
Abstract
Unexpected drug–drug interactions (DDIs) are important issues for both pharmaceutical research and clinical applications due to the high risk of causing severe adverse drug reactions or drug withdrawals. Many deep learning models have achieved high performance in DDI prediction, but model interpretability to reveal the underlying causes of DDIs has not been extensively explored. Here we propose MeTDDI—a deep learning framework with local–global self-attention and co-attention to learn motif-based graphs for DDI prediction. MeTDDI achieved competitive performance compared with state-of-the-art models. Regarding interpretability, we conducted extensive assessments on 73 drugs with 13,786 DDIs and MeTDDI can precisely explain the structural mechanisms for 5,602 DDIs involving 58 drugs. Besides, MeTDDI shows potential to explain complex DDI mechanisms and mitigate DDI risks. To summarize, MeTDDI provides a new perspective on exploring DDI mechanisms, which will benefit both drug discovery and polypharmacy for safer therapies for patients. A transformer-based approach that predicts drug–drug interactions in polypharmacy has been shown, which also identifies perpetrator drugs and the chemical mechanisms causing the interactions. The method could facilitate high-throughput optimization of drug combinations and mitigate adverse drug–drug interaction risks.
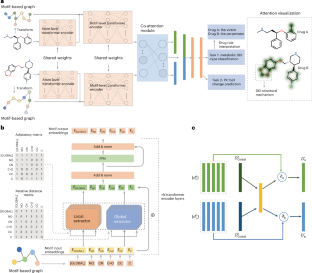
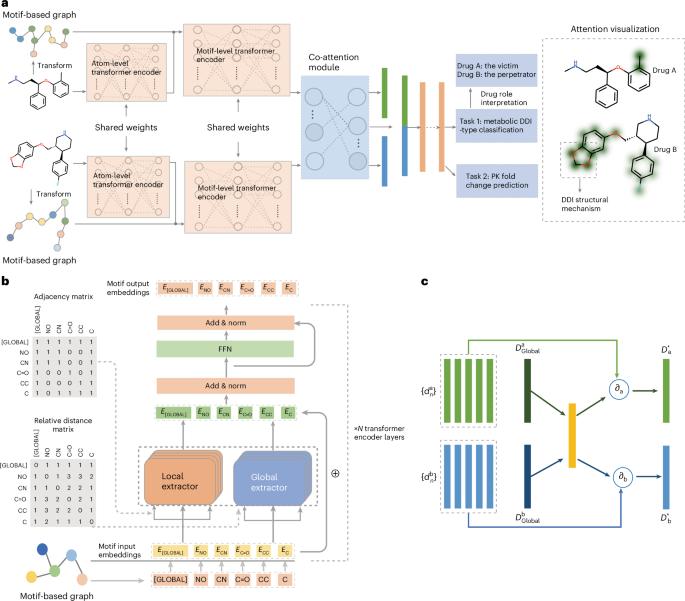
通过局部-全局自我关注,学习基于图案的药物相互作用预测图谱
意外的药物相互作用(DDIs)是药物研究和临床应用中的重要问题,因为它极有可能导致严重的药物不良反应或停药。许多深度学习模型在 DDI 预测方面取得了很高的性能,但揭示 DDIs 潜在原因的模型可解释性尚未得到广泛探索。在此,我们提出了MeTDDI--一种具有局部-全局自关注和共关注的深度学习框架,用于学习基于主题图的DDI预测。与最先进的模型相比,MeTDDI 的性能极具竞争力。在可解释性方面,我们对73种药物的13786个DDIs进行了广泛评估,MeTDDI可以精确解释58种药物的5602个DDIs的结构机制。此外,MeTDDI 显示出解释复杂 DDI 机制和降低 DDI 风险的潜力。总之,MeTDDI 为探索 DDI 机制提供了一个新的视角,这将有利于药物发现和多药治疗,为患者提供更安全的疗法。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Nature Machine Intelligence
Multiple-
CiteScore
36.90
自引率
2.10%
发文量
127
期刊介绍:
Nature Machine Intelligence is a distinguished publication that presents original research and reviews on various topics in machine learning, robotics, and AI. Our focus extends beyond these fields, exploring their profound impact on other scientific disciplines, as well as societal and industrial aspects. We recognize limitless possibilities wherein machine intelligence can augment human capabilities and knowledge in domains like scientific exploration, healthcare, medical diagnostics, and the creation of safe and sustainable cities, transportation, and agriculture. Simultaneously, we acknowledge the emergence of ethical, social, and legal concerns due to the rapid pace of advancements.
To foster interdisciplinary discussions on these far-reaching implications, Nature Machine Intelligence serves as a platform for dialogue facilitated through Comments, News Features, News & Views articles, and Correspondence. Our goal is to encourage a comprehensive examination of these subjects.
Similar to all Nature-branded journals, Nature Machine Intelligence operates under the guidance of a team of skilled editors. We adhere to a fair and rigorous peer-review process, ensuring high standards of copy-editing and production, swift publication, and editorial independence.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


