A deep learning method that identifies cellular heterogeneity using nanoscale nuclear features
IF 18.8
1区 计算机科学
Q1 COMPUTER SCIENCE, ARTIFICIAL INTELLIGENCE
引用次数: 0
Abstract
Cellular phenotypic heterogeneity is an important hallmark of many biological processes and understanding its origins remains a substantial challenge. This heterogeneity often reflects variations in the chromatin structure, influenced by factors such as viral infections and cancer, which dramatically reshape the cellular landscape. To address the challenge of identifying distinct cell states, we developed artificial intelligence of the nucleus (AINU), a deep learning method that can identify specific nuclear signatures at the nanoscale resolution. AINU can distinguish different cell states based on the spatial arrangement of core histone H3, RNA polymerase II or DNA from super-resolution microscopy images. With only a small number of images as the training data, AINU correctly identifies human somatic cells, human-induced pluripotent stem cells, very early stage infected cells transduced with DNA herpes simplex virus type 1 and even cancer cells after appropriate retraining. Finally, using AI interpretability methods, we find that the RNA polymerase II localizations in the nucleoli aid in distinguishing human-induced pluripotent stem cells from their somatic cells. Overall, AINU coupled with super-resolution microscopy of nuclear structures provides a robust tool for the precise detection of cellular heterogeneity, with considerable potential for advancing diagnostics and therapies in regenerative medicine, virology and cancer biology. Cellular phenotypic heterogeneity is a key determinant of biological functions and is challenging to identify. A deep learning method that recognizes specific nuclear signatures is discussed, which can identify cellular heterogeneity and differentiate between various cell states using a small amount of super-resolution microscopy data.
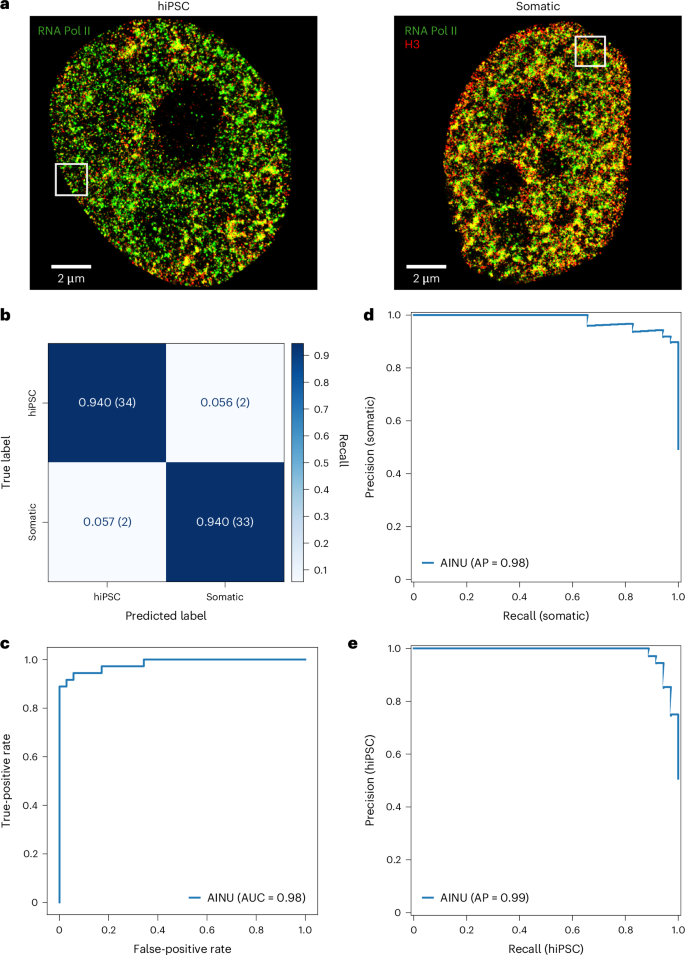
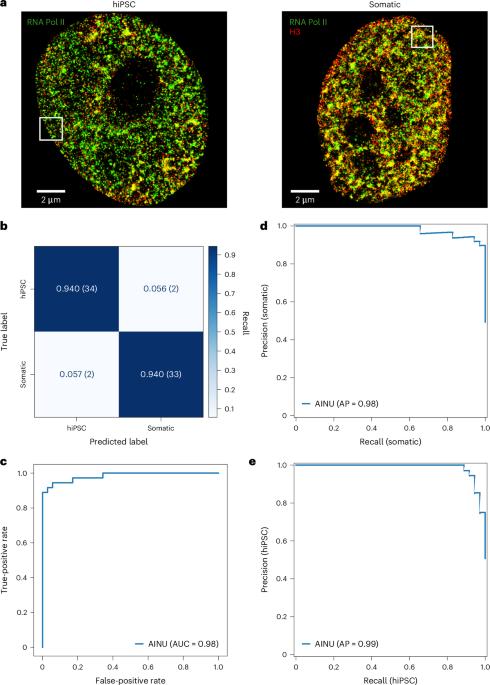
利用纳米级核特征识别细胞异质性的深度学习方法
细胞表型异质性是许多生物过程的重要标志,而了解其起源仍是一项巨大的挑战。这种异质性通常反映了染色质结构的变化,受病毒感染和癌症等因素的影响,这些因素极大地重塑了细胞景观。为了应对识别不同细胞状态的挑战,我们开发了细胞核人工智能(AINU),这是一种深度学习方法,可以在纳米级分辨率上识别特定的核特征。AINU 可以根据超分辨率显微镜图像中核心组蛋白 H3、RNA 聚合酶 II 或 DNA 的空间排列来区分不同的细胞状态。AINU 仅用少量图像作为训练数据,就能正确识别人类体细胞、人类诱导多能干细胞、DNA 1 型单纯疱疹病毒转导的早期感染细胞,甚至在经过适当的再训练后还能识别癌细胞。最后,利用人工智能可解释性方法,我们发现核小体中 RNA 聚合酶 II 的定位有助于区分人类诱导多能干细胞和体细胞。总之,AINU 与核结构超分辨率显微镜相结合,为精确检测细胞异质性提供了一个强大的工具,在再生医学、病毒学和癌症生物学的诊断和治疗方面具有相当大的潜力。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Nature Machine Intelligence
Multiple-
CiteScore
36.90
自引率
2.10%
发文量
127
期刊介绍:
Nature Machine Intelligence is a distinguished publication that presents original research and reviews on various topics in machine learning, robotics, and AI. Our focus extends beyond these fields, exploring their profound impact on other scientific disciplines, as well as societal and industrial aspects. We recognize limitless possibilities wherein machine intelligence can augment human capabilities and knowledge in domains like scientific exploration, healthcare, medical diagnostics, and the creation of safe and sustainable cities, transportation, and agriculture. Simultaneously, we acknowledge the emergence of ethical, social, and legal concerns due to the rapid pace of advancements.
To foster interdisciplinary discussions on these far-reaching implications, Nature Machine Intelligence serves as a platform for dialogue facilitated through Comments, News Features, News & Views articles, and Correspondence. Our goal is to encourage a comprehensive examination of these subjects.
Similar to all Nature-branded journals, Nature Machine Intelligence operates under the guidance of a team of skilled editors. We adhere to a fair and rigorous peer-review process, ensuring high standards of copy-editing and production, swift publication, and editorial independence.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


