Extreme phenotype sampling and next generation sequencing to identify genetic variants associated with tacrolimus in African American kidney transplant recipients
IF 2.9
3区 医学
Q2 GENETICS & HEREDITY
引用次数: 0
Abstract
African American (AA) kidney transplant recipients (KTRs) have poor outcomes, which may in-part be due to tacrolimus (TAC) sub-optimal immunosuppression. We previously determined the common genetic regulators of TAC pharmacokinetics in AAs which were CYP3A5 *3, *6, and *7. To identify low-frequency variants that impact TAC pharmacokinetics, we used extreme phenotype sampling and compared individuals with extreme high (n = 58) and low (n = 60) TAC troughs (N = 515 AA KTRs). Targeted next generation sequencing was conducted in these two groups. Median TAC troughs in the high group were 7.7 ng/ml compared with 6.3 ng/ml in the low group, despite lower daily doses of 5 versus 12 mg, respectively. Of 34,542 identified variants across 99 genes, 1406 variants were suggestively associated with TAC troughs in univariate models (p-value < 0.05), however none were significant after multiple testing correction. We suggest future studies investigate additional sources of TAC pharmacokinetic variability such as drug-drug-gene interactions and pharmacomicrobiome.
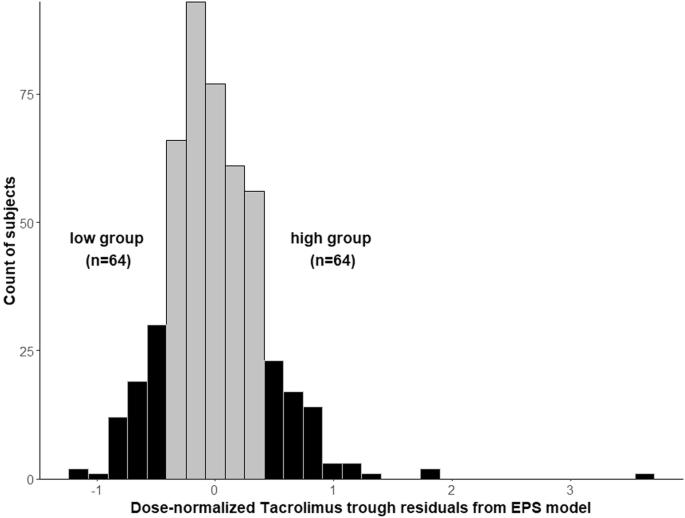
通过极端表型取样和新一代测序技术,确定非裔美国肾移植受者中与他克莫司相关的基因变异。
非裔美国人(AA)肾移植受者(KTR)的治疗效果不佳,部分原因可能是他克莫司(TAC)的免疫抑制效果不理想。我们之前确定了 AAs 中 TAC 药代动力学的常见遗传调节因子,即 CYP3A5 *3、*6 和 *7。为了确定影响 TAC 药代动力学的低频变异,我们使用了极端表型取样,并比较了 TAC 谷值极高(n = 58)和极低(n = 60)的个体(N = 515 AA KTR)。在这两组人中进行了有针对性的新一代测序。尽管每日剂量分别为 5 毫克和 12 毫克,但高剂量组的中位 TAC 谷值为 7.7 纳克/毫升,而低剂量组为 6.3 纳克/毫升。在 99 个基因的 34,542 个已鉴定变异中,有 1406 个变异在单变量模型中与 TAC 谷值有提示性关联(p 值为 0.5)。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Pharmacogenomics Journal
医学-药学
CiteScore
7.20
自引率
0.00%
发文量
35
审稿时长
6-12 weeks
期刊介绍:
The Pharmacogenomics Journal is a print and electronic journal, which is dedicated to the rapid publication of original research on pharmacogenomics and its clinical applications.
Key areas of coverage include:
Personalized medicine
Effects of genetic variability on drug toxicity and efficacy
Identification and functional characterization of polymorphisms relevant to drug action
Pharmacodynamic and pharmacokinetic variations and drug efficacy
Integration of new developments in the genome project and proteomics into clinical medicine, pharmacology, and therapeutics
Clinical applications of genomic science
Identification of novel genomic targets for drug development
Potential benefits of pharmacogenomics.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


