Expression and prognostic value of cell-cycle-associated genes in lung squamous cell carcinoma
Abstract
Background
Lung cancer continues to be a prevalent cause of cancer-related deaths worldwide, with lung squamous carcinoma (LUSC) being a significant subtype characterized by comparatively low survival rates. Extensive molecular studies on LUSC have been conducted; however, the clinical importance of cell-cycle-associated genes has rarely been examined. This study aimed to investigate the relationship between these genes and LUSC.
Methods
The expression trends of genes related to the cell cycle in a group of patients with LUSC were analyzed. Clinical information and mRNA expression data were obtained from The Cancer Genome Atlas via cBioportal. Multiple analyses have been performed to investigate the association between these genes and LUSC.
Results
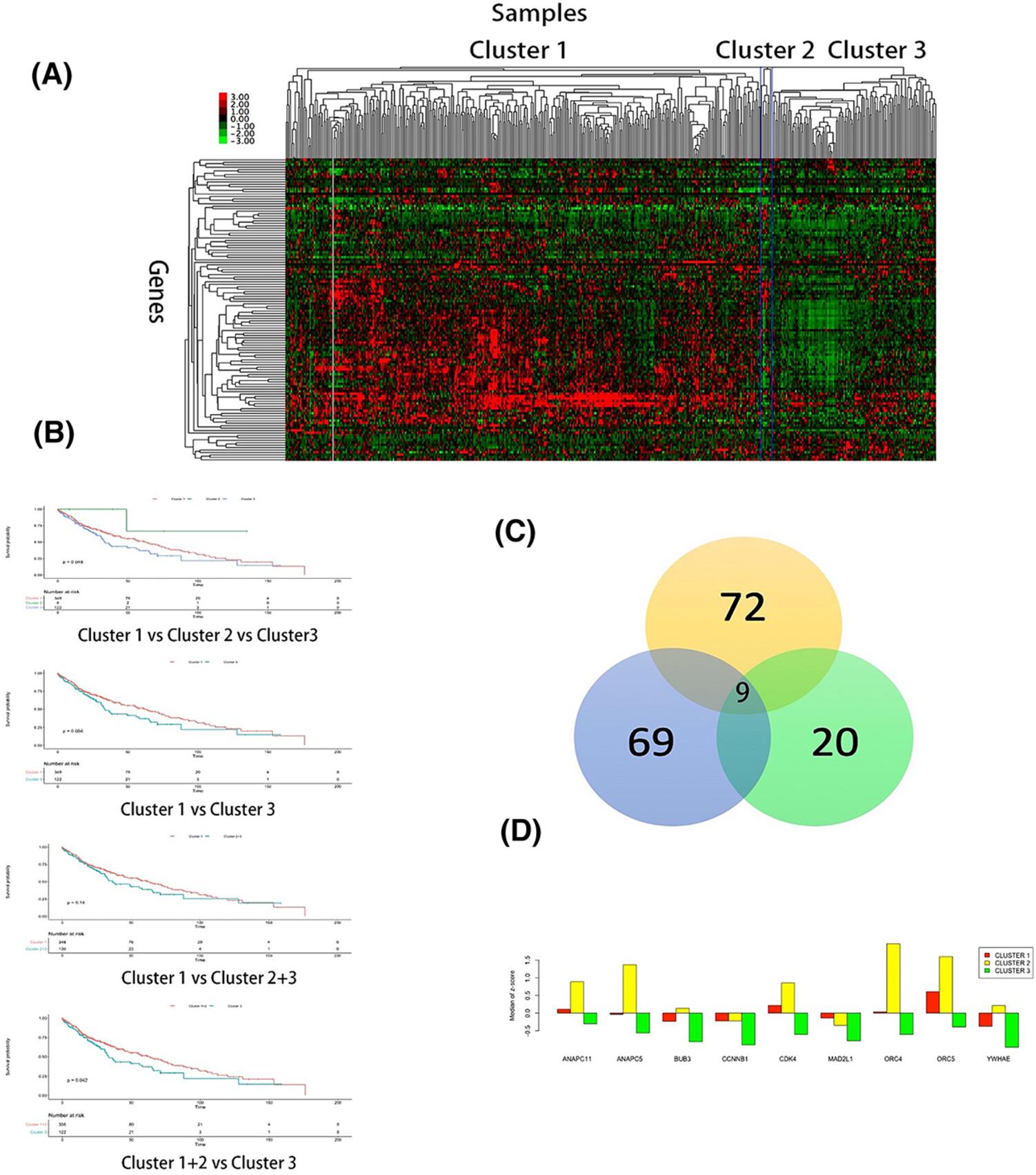
Three clusters were identified based on the mRNA expression of 124 cell cycle-associated genes. Cluster 3 exhibited the worst prognosis. A comparative analysis showed that nine expressed genes differed distinctly among all the clusters. Among these nine genes, elevated expression of CDK4 was strongly associated with positive prognosis. Furthermore, the expression of ANAPC11, ANAPC5, and ORC4 correlated with the advancement of LUSC pathological stages.
Conclusions
Gene expression profiles associated with the cell cycle across various LUSC subtypes were identified, highlighting that specific genes are related to prognosis and disease stages. Based on these results, new prognostic strategies, patient stratification, and targeted therapy trials have been conducted for LUSC.

 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


