Chromosome-contiguous genome for the Haecon-5 strain of Haemonchus contortus reveals marked genetic variability and enables the discovery of essential gene candidates
IF 3.7
2区 医学
Q1 PARASITOLOGY
引用次数: 0
Abstract
Millions of livestock animals worldwide are infected with the haematophagous barber’s pole worm, Haemonchus contortus, the aetiological agent of haemonchosis. Despite the major significance of this parasite worldwide and its widespread resistance to current treatments, the lack of a high-quality genome for the well-defined strain of this parasite from Australia, called Haecon-5, has constrained research in a number of areas including host-parasite interactions, drug discovery and population genetics. To enable research in these areas, we report here a chromosome-contiguous genome (∼280 Mb) for Haecon-5 with high-quality models for 19,234 protein-coding genes. Comparative genomic analyses show significant genomic similarity (synteny) with a UK strain of H. contortus, called MHco3(ISE).N1 (abbreviated as “ISE”), but we also discover marked differences in genomic structure/gene arrangements, distribution of nucleotide variability (single nucleotide polymorphisms (SNPs) and indels) and orthology between Haecon-5 and ISE. We used the genome and extensive transcriptomic resources for Haecon-5 to predict a subset of essential single-copy genes employing a “cross-species” machine learning (ML) approach using a range of features from nucleotide/protein sequences, protein orthology, subcellular localisation, single-cell RNA-seq and/or histone methylation data available for the model organisms Caenorhabditis elegans and Drosophila melanogaster. From a set of 1,464 conserved single copy genes, transcribed in key life-cycle stages of H. contortus, we identified 232 genes whose homologs have critical functions in C. elegans and/or D. melanogaster, and prioritised 10 of them for further characterisation; nine of the 10 genes likely play roles in neurophysiological processes, germline, hypodermis and/or respiration, and one is an unknown (orphan) gene for which no detailed functional information exists. Future studies of these genes/gene products are warranted to elucidate their roles in parasite biology, host-parasite interplay and/or disease. Clearly, the present Haecon-5 reference genome and associated resources now underpin a broad range of fundamental investigations of H. contortus and could assist in accelerating the discovery of novel intervention targets and drug candidates to combat haemonchosis.
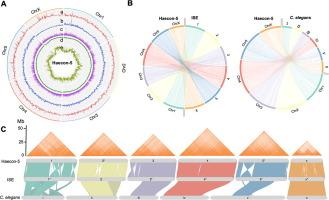
Haecon-5血吸虫菌株的染色体连续基因组揭示了明显的遗传变异,有助于发现重要的候选基因。
全世界有数以百万计的牲畜感染了噬血性理发杆蠕虫(Haemonchus contortus),这是血吸虫病的病原体。尽管这种寄生虫在世界范围内具有重要意义,而且对目前的治疗方法普遍存在抗药性,但由于缺乏澳大利亚寄生虫明确菌株(Haecon-5)的高质量基因组,制约了宿主与寄生虫相互作用、药物发现和种群遗传学等多个领域的研究。为了促进这些领域的研究,我们在此报告了 Haecon-5 的染色体连续基因组(∼280 Mb),其中包含 19,234 个蛋白质编码基因的高质量模型。比较基因组分析表明,Haecon-5 与英国的一个 H. contortus 株系 MHco3(ISE).N1(缩写为 "ISE")在基因组结构/基因排列、核苷酸变异性(单核苷酸多态性(SNP)和嵌合体)分布和选系方面存在显著的相似性(同源),但我们也发现 Haecon-5 与 ISE 在基因组结构/基因排列、核苷酸变异性(单核苷酸多态性(SNP)和嵌合体)分布和选系方面存在显著的差异。我们利用 Haecon-5 的基因组和广泛的转录组资源,采用 "跨物种 "机器学习(ML)方法,利用模式生物秀丽隐杆线虫(Caenorhabditis elegans)和黑腹果蝇(Drosophila melanogaster)的核苷酸/蛋白质序列、蛋白质同源、亚细胞定位、单细胞 RNA-seq 和/或组蛋白甲基化数据等一系列特征,预测了基本单拷贝基因的子集。在一组 1464 个保守的单拷贝基因中,我们确定了 232 个在秀丽隐杆线虫和/或黑腹果蝇中具有关键功能的同源基因,并优先对其中 10 个基因进行了进一步鉴定;这 10 个基因中的 9 个可能在神经生理过程、生殖细胞、下胚层和/或呼吸中发挥作用,还有一个是未知(孤儿)基因,没有详细的功能信息。今后有必要对这些基因/基因产物进行研究,以阐明它们在寄生虫生物学、宿主-寄生虫相互作用和/或疾病中的作用。很明显,目前的 Haecon-5 参考基因组及相关资源是对霍乱弧菌进行广泛基础研究的基础,并有助于加快发现新的干预目标和候选药物,以防治霍乱弧菌病。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊
CiteScore
8.40
自引率
2.50%
发文量
76
审稿时长
23 days
期刊介绍:
International Journal for Parasitology offers authors the option to sponsor nonsubscriber access to their articles on Elsevier electronic publishing platforms. For more information please view our Sponsored Articles page. The International Journal for Parasitology publishes the results of original research in all aspects of basic and applied parasitology, including all the fields covered by its Specialist Editors, and ranging from parasites and host-parasite relationships of intrinsic biological interest to those of social and economic importance in human and veterinary medicine and agriculture.

 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


