Building endoplasmic reticulum stress-related LncRNAs signatures of lung adenocarcinoma
Abstract
Background
Endoplasmic reticulum stress (ERS) could be a strategy for treating malignant tumors. Moreover, long noncoding RNAs (lncRNAs) can promote tumorigenesis and progression, and forecast the prognosis of cancers. Nevertheless, the prognostic value of ERS-related lncRNAs has not been reported in lung adenocarcinoma (LUAD).
Methods
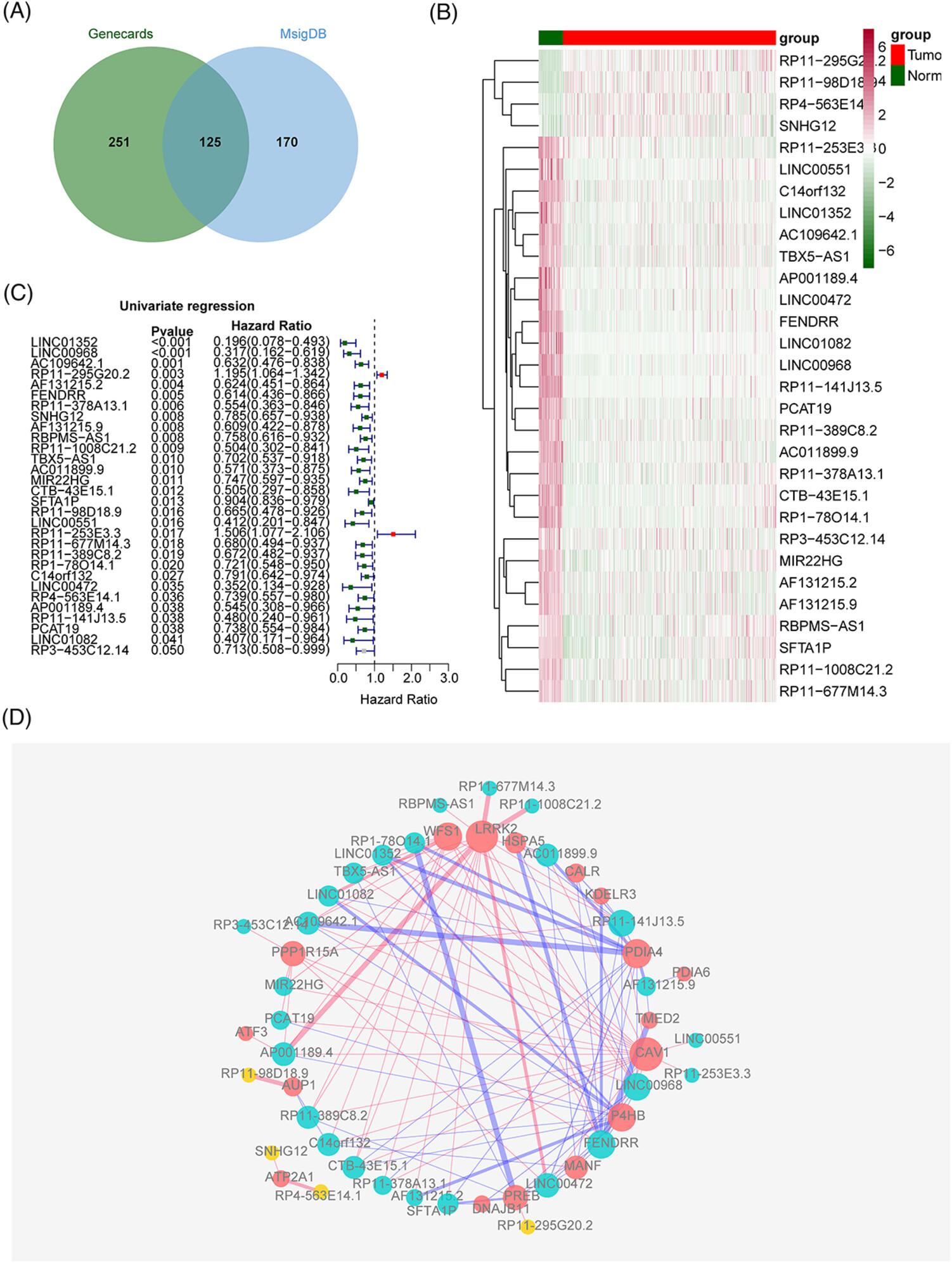
The messenger RNA (mRNA), microRNA (miRNA) and lncRNA expression data related to LUAD were obtained in public databases (TCGA and GEO databases). Prognostic ERS-related differentially expressed lncRNAs (ERS-DELs) were obtained and used to build an ERS-related model by Cox regression analysis. Moreover, we further screened independent prognostic elements and built a nomogram. Furthermore, enrichment analysis of genes was conducted to investigate the functions. A lncRNA–miRNA–mRNA network was built to explore mechanism of lncRNAs. Finally, qRT-PCR was utilized to examine the expression levels of lncRNAs.
Results
30 ERS-DELs were identified, and an ERS-related signature was built based on AF131215.2, LINC00472, LINC01352, RP1-78O14.1, RP11-253E3.3, RP11-98D18.9, and SNHG12. Gene set enrichment analysis indicated that genes in the high-risk group were chiefly focused on the regulation of mRNA binding, and genes in the low-risk group were significantly focused on protein localization to cilia. A lncRNA–miRNA–mRNA network, containing 7 signature lncRNAs, 23 miRNAs, and 128 mRNAs, was also established. Eventually, quantitative real-time polymerase chain reaction was used to confirm that seven prognostic lncRNAs had a consistent expression with the analysis.
Conclusions
An ERS-related signature containing seven prognostic lncRNAs was built, which offered new thinking concerning the role of ERS-related lncRNAs in LUAD.

 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


