Spatial proteomic profiling elucidates immune determinants of neoadjuvant chemo-immunotherapy in esophageal squamous cell carcinoma
IF 6.9
1区 医学
Q1 BIOCHEMISTRY & MOLECULAR BIOLOGY
引用次数: 0
Abstract
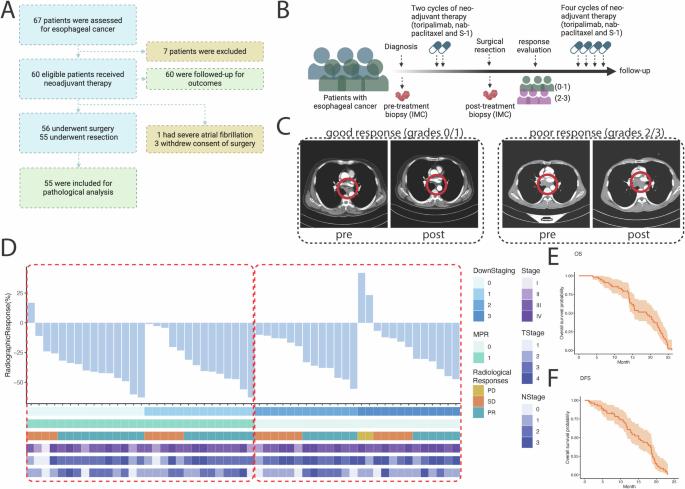
Esophageal squamous cell carcinoma (ESCC) presents significant clinical and therapeutic challenges due to its aggressive nature and generally poor prognosis. We initiated a Phase II clinical trial (ChiCTR1900027160) to assess the efficacy of a pioneering neoadjuvant chemo-immunotherapy regimen comprising programmed death-1 (PD-1) blockade (Toripalimab), nanoparticle albumin-bound paclitaxel (nab-paclitaxel), and the oral fluoropyrimidine derivative S-1, in patients with locally advanced ESCC. This study uniquely integrates clinical outcomes with advanced spatial proteomic profiling using Imaging Mass Cytometry (IMC) to elucidate the dynamics within the tumor microenvironment (TME), focusing on the mechanistic interplay of resistance and response. Sixty patients participated, receiving the combination therapy prior to surgical resection. Our findings demonstrated a major pathological response (MPR) in 62% of patients and a pathological complete response (pCR) in 29%. The IMC analysis provided a detailed regional assessment, revealing that the spatial arrangement of immune cells, particularly CD8+ T cells and B cells within tertiary lymphoid structures (TLS), and S100A9+ inflammatory macrophages in fibrotic regions are predictive of therapeutic outcomes. Employing machine learning approaches, such as support vector machine (SVM) and random forest (RF) analysis, we identified critical spatial features linked to drug resistance and developed predictive models for drug response, achieving an area under the curve (AUC) of 97%. These insights underscore the vital role of integrating spatial proteomics into clinical trials to dissect TME dynamics thoroughly, paving the way for personalized and precise cancer treatment strategies in ESCC. This holistic approach not only enhances our understanding of the mechanistic basis behind drug resistance but also sets a robust foundation for optimizing therapeutic interventions in ESCC.
空间蛋白质组剖析阐明食管鳞状细胞癌新辅助化疗免疫疗法的免疫决定因素
食管鳞状细胞癌(ESCC)因其侵袭性强、预后普遍较差,给临床和治疗带来了巨大挑战。我们启动了一项 II 期临床试验(ChiCTR1900027160),评估由程序性死亡-1(PD-1)阻断剂(托里帕利单抗)、纳米颗粒白蛋白结合型紫杉醇(nab-紫杉醇)和口服氟嘧啶衍生物 S-1 组成的开创性新辅助化疗免疫疗法方案对局部晚期 ESCC 患者的疗效。这项研究独特地将临床结果与使用成像质控仪(IMC)进行的高级空间蛋白质组分析相结合,以阐明肿瘤微环境(TME)内的动态变化,重点研究耐药性和反应的机理相互作用。60名患者在手术切除前接受了联合疗法。我们的研究结果显示,62%的患者获得了主要病理反应(MPR),29%的患者获得了病理完全反应(pCR)。IMC分析提供了详细的区域评估,揭示了免疫细胞的空间排列,尤其是三级淋巴结构(TLS)中的CD8+ T细胞和B细胞,以及纤维化区域中的S100A9+炎性巨噬细胞对治疗结果的预测作用。利用支持向量机(SVM)和随机森林(RF)分析等机器学习方法,我们确定了与耐药性相关的关键空间特征,并建立了药物反应预测模型,其曲线下面积(AUC)达到了 97%。这些见解凸显了将空间蛋白质组学融入临床试验以彻底剖析TME动态的重要作用,从而为ESCC的个性化精准癌症治疗策略铺平了道路。这种全面的方法不仅增强了我们对耐药性背后机理基础的了解,还为优化 ESCC 的治疗干预奠定了坚实的基础。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Oncogene
医学-生化与分子生物学
CiteScore
15.30
自引率
1.20%
发文量
404
审稿时长
1 months
期刊介绍:
Oncogene is dedicated to advancing our understanding of cancer processes through the publication of exceptional research. The journal seeks to disseminate work that challenges conventional theories and contributes to establishing new paradigms in the etio-pathogenesis, diagnosis, treatment, or prevention of cancers. Emphasis is placed on research shedding light on processes driving metastatic spread and providing crucial insights into cancer biology beyond existing knowledge.
Areas covered include the cellular and molecular biology of cancer, resistance to cancer therapies, and the development of improved approaches to enhance survival. Oncogene spans the spectrum of cancer biology, from fundamental and theoretical work to translational, applied, and clinical research, including early and late Phase clinical trials, particularly those with biologic and translational endpoints.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


