Deep learning prediction of glycopeptide tandem mass spectra powers glycoproteomics
IF 18.8
1区 计算机科学
Q1 COMPUTER SCIENCE, ARTIFICIAL INTELLIGENCE
引用次数: 0
Abstract
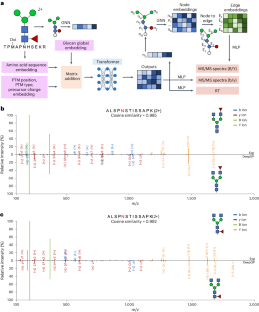
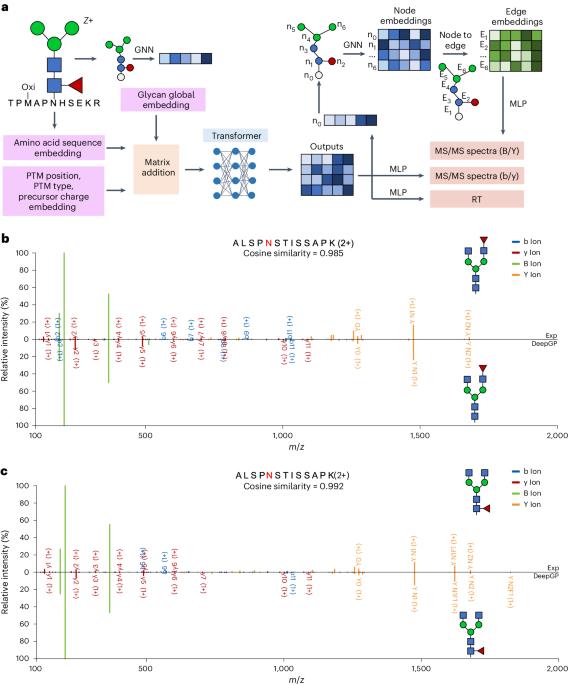
Protein glycosylation, a post-translational modification of proteins by glycans, plays an important role in numerous physiological and pathological cellular functions. Glycoproteomics, the study of protein glycosylation on a proteome-wide scale, utilizes liquid chromatography coupled with tandem mass spectrometry (MS/MS) to get combinational information on glycosylation site, glycosylation level and glycan structure. However, current database searching methods for glycoproteomics often struggle with glycan structure determination due to the limited occurrence of structure-determining ions. Although spectral searching methods can leverage fragment intensity to facilitate the structure identification of glycopeptides, their application is hindered by difficulties in spectral library construction. In this work, we present DeepGP, a hybrid deep learning framework based on transformer and graph neural networks, for the prediction of MS/MS spectra and retention time of glycopeptides. Two graph neural network modules are employed to capture the branched glycan structure and predict glycan ion intensity, respectively. Additionally, a pretraining strategy is implemented to alleviate the insufficiency of glycoproteomics data. Testing on multiple biological datasets, DeepGP accurately predicts MS/MS spectra and retention time of glycopeptides, closely aligning with the experimental results. Comprehensive benchmarking of DeepGP on synthetic and biological datasets validates its effectiveness in distinguishing similar glycans. Based on various decoy methods, DeepGP in combination with database searching can increase glycopeptide detection sensitivity. We anticipate that DeepGP can inspire extensive future work in glycoproteomics. Glycosylation, a prevalent type of post-translational modification of proteins by glycan molecules, plays a major role in the proteome. Zong et al. present DeepGP, a hybrid deep learning framework based on transformer and graph neural network architectures that accurately predicts tandem mass spectra and retention times of glycopeptides, providing information on glycosylation and glycan structure.
糖肽串联质谱的深度学习预测为糖蛋白组学提供动力
蛋白质糖基化是蛋白质通过聚糖进行的翻译后修饰,在细胞的多种生理和病理功能中发挥着重要作用。糖蛋白组学是在整个蛋白质组范围内研究蛋白质糖基化的方法,它利用液相色谱法和串联质谱法(MS/MS)获得糖基化位点、糖基化水平和聚糖结构的综合信息。然而,目前用于糖蛋白组学的数据库搜索方法往往由于结构决定离子的出现有限而难以确定糖分子结构。虽然光谱搜索方法可以利用片段强度来促进糖肽的结构鉴定,但其应用受到光谱库构建困难的阻碍。在这项工作中,我们提出了基于变换器和图神经网络的混合深度学习框架 DeepGP,用于预测糖肽的 MS/MS 图谱和保留时间。我们采用了两个图神经网络模块,分别用于捕捉支链聚糖结构和预测聚糖离子强度。此外,还采用了预训练策略,以缓解糖蛋白组学数据不足的问题。在多个生物数据集上进行测试后,DeepGP 准确预测了糖肽的 MS/MS 图谱和保留时间,与实验结果非常吻合。在合成和生物数据集上对 DeepGP 进行的全面基准测试验证了它在区分相似聚糖方面的有效性。基于各种诱饵方法,DeepGP 与数据库搜索相结合可以提高糖肽检测灵敏度。我们预计,DeepGP 将激发未来在糖蛋白组学领域的广泛工作。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Nature Machine Intelligence
Multiple-
CiteScore
36.90
自引率
2.10%
发文量
127
期刊介绍:
Nature Machine Intelligence is a distinguished publication that presents original research and reviews on various topics in machine learning, robotics, and AI. Our focus extends beyond these fields, exploring their profound impact on other scientific disciplines, as well as societal and industrial aspects. We recognize limitless possibilities wherein machine intelligence can augment human capabilities and knowledge in domains like scientific exploration, healthcare, medical diagnostics, and the creation of safe and sustainable cities, transportation, and agriculture. Simultaneously, we acknowledge the emergence of ethical, social, and legal concerns due to the rapid pace of advancements.
To foster interdisciplinary discussions on these far-reaching implications, Nature Machine Intelligence serves as a platform for dialogue facilitated through Comments, News Features, News & Views articles, and Correspondence. Our goal is to encourage a comprehensive examination of these subjects.
Similar to all Nature-branded journals, Nature Machine Intelligence operates under the guidance of a team of skilled editors. We adhere to a fair and rigorous peer-review process, ensuring high standards of copy-editing and production, swift publication, and editorial independence.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


