Experimental confirmation and bioinformatics reveal biomarkers of immune system infiltration and hypertrophy ligamentum flavum
Abstract
Background
Hypertrophy ligamentum flavum is a prevalent chronic spinal condition that affects middle-aged and older adults. However, the molecular pathways behind this disease are not well comprehended.
Objective
The objective of this work is to implement bioinformatics techniques in order to identify crucial biological markers and immune infiltration that are linked to hypertrophy ligamentum flavum. Further, the study aims to experimentally confirm the molecular mechanisms that underlie the hypertrophy ligamentum flavum.
Methods
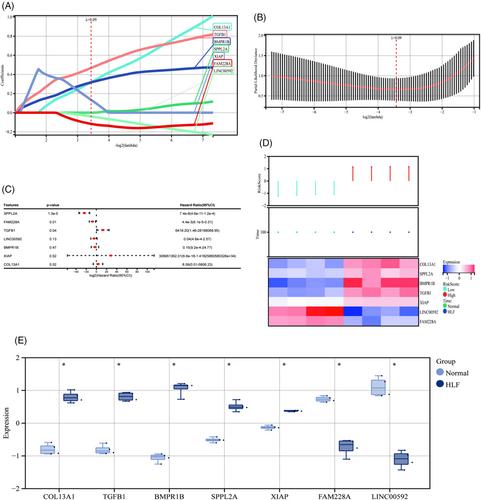
The corresponding gene expression profiles (GSE113212) were selected from a comprehensive gene expression database. The gene dataset for hypertrophy ligamentum flavum was acquired from GeneCards. A network of interactions between proteins was created, and an analysis of functional enrichment was conducted using the Kyoto Encyclopedia of Genes and Genomes (KEGG) and Gene Ontology (GO) databases. An study of hub genes was performed to evaluate the infiltration of immune cells in patient samples compared to tissues from the control group. Finally, samples of the ligamentum flavum were taken with the purpose of validating the expression of important genes in a clinical setting.
Results
Overall, 27 hub genes that were differently expressed were found through molecular biology. The hub genes were found to be enriched in immune response, chemokine-mediated signaling pathways, inflammation, ossification, and fibrosis processes, as demonstrated by GO and KEGG studies. The main signaling pathways involved include the TNF signaling pathway, cytokine–cytokine receptor interaction, and TGF-β signaling pathway. An examination of immunocell infiltration showed notable disparities in B cells (naïve and memory) and activated T cells (CD4 memory) between patients with hypertrophic ligamentum flavum and the control group of healthy individuals. The in vitro validation revealed markedly elevated levels of ossification and fibrosis-related components in the hypertrophy ligamentum flavum group, as compared to the normal group.
Conclusion
The TGF-β signaling pathway, TNF signaling pathway, and related hub genes play crucial roles in the progression of ligamentum flavum hypertrophic. Our study may guide future research on fibrosis of the ligamentum flavum.

 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


