Sequential Patching Lattice for Image Classification and Enquiry
IF 4.7
2区 医学
Q1 PATHOLOGY
引用次数: 0
Abstract
Digital pathology and the integration of artificial intelligence (AI) models have revolutionized histopathology, opening new opportunities. With the increasing availability of whole-slide images (WSIs), demand is growing for efficient retrieval, processing, and analysis of relevant images from vast biomedical archives. However, processing WSIs presents challenges due to their large size and content complexity. Full computer digestion of WSIs is impractical, and processing all patches individually is prohibitively expensive. In this article, we propose an unsupervised patching algorithm, Sequential Patching Lattice for Image Classification and Enquiry (SPLICE). This novel approach condenses a histopathology WSI into a compact set of representative patches, forming a collage of WSI while minimizing redundancy. SPLICE prioritizes patch quality and uniqueness by sequentially analyzing a WSI and selecting nonredundant representative features. In search and match applications, SPLICE showed improved accuracy, reduced computation time, and storage requirements compared with existing state-of-the-art methods. As an unsupervised method, SPLICE effectively reduced storage requirements for representing tissue images by 50%. This reduction can enable numerous algorithms in computational pathology to operate much more efficiently, paving the way for accelerated adoption of digital pathology.
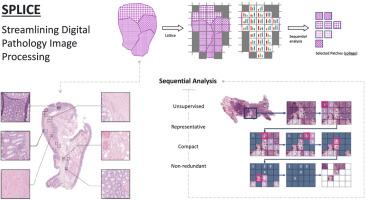
SPLICE - 简化数字病理图像处理。
数字病理学和人工智能(AI)模型的整合彻底改变了组织病理学,带来了新的机遇。随着整张切片图像(WSI)的可用性不断提高,从庞大的生物医学档案中高效检索、处理和分析相关图像的需求也日益增长。然而,由于 WSIs 体积庞大、内容复杂,处理 WSIs 是一项挑战。完全用计算机消化 WSI 是不切实际的,而单独处理所有补丁又过于昂贵。在本文中,我们提出了一种无监督修补算法--用于图像分类和查询的序列修补网格(SPLICE)。这种新方法将组织病理学 WSI 浓缩为一组紧凑的代表性补丁,形成 WSI 的 "拼贴",同时最大限度地减少冗余。SPLICE 通过顺序分析 WSI 并选择非冗余的代表性特征,优先考虑斑块的质量和独特性。我们对 SPLICE 的搜索和匹配应用进行了评估,结果表明,与现有的先进方法相比,SPLICE 提高了准确性,减少了计算时间和存储要求。作为一种无监督方法,SPLICE 能有效地将表示组织图像的存储要求降低 50%。这种减少使计算病理学中的许多算法能更有效地运行,为加快数字病理学的应用铺平了道路。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊
CiteScore
11.40
自引率
0.00%
发文量
178
审稿时长
30 days
期刊介绍:
The American Journal of Pathology, official journal of the American Society for Investigative Pathology, published by Elsevier, Inc., seeks high-quality original research reports, reviews, and commentaries related to the molecular and cellular basis of disease. The editors will consider basic, translational, and clinical investigations that directly address mechanisms of pathogenesis or provide a foundation for future mechanistic inquiries. Examples of such foundational investigations include data mining, identification of biomarkers, molecular pathology, and discovery research. Foundational studies that incorporate deep learning and artificial intelligence are also welcome. High priority is given to studies of human disease and relevant experimental models using molecular, cellular, and organismal approaches.

 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


