Reinforcement learning-based anatomical maps for pancreas subregion and duct segmentation
Abstract
Background
The pancreas is a complex abdominal organ with many anatomical variations, and therefore automated pancreas segmentation from medical images is a challenging application.
Purpose
In this paper, we present a framework for segmenting individual pancreatic subregions and the pancreatic duct from three-dimensional (3D) computed tomography (CT) images.
Methods
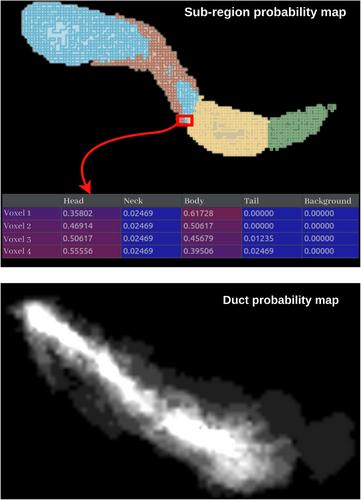
A multiagent reinforcement learning (RL) network was used to detect landmarks of the head, neck, body, and tail of the pancreas, and landmarks along the pancreatic duct in a selected target CT image. Using the landmark detection results, an atlas of pancreases was nonrigidly registered to the target image, resulting in anatomical probability maps for the pancreatic subregions and duct. The probability maps were augmented with multilabel 3D U-Net architectures to obtain the final segmentation results.
Results
To evaluate the performance of our proposed framework, we computed the Dice similarity coefficient (DSC) between the predicted and ground truth manual segmentations on a database of 82 CT images with manually segmented pancreatic subregions and 37 CT images with manually segmented pancreatic ducts. For the four pancreatic subregions, the mean DSC improved from 0.38, 0.44, and 0.39 with standard 3D U-Net, Attention U-Net, and shifted windowing (Swin) U-Net architectures, to 0.51, 0.47, and 0.49, respectively, when utilizing the proposed RL-based framework. For the pancreatic duct, the RL-based framework achieved a mean DSC of 0.70, significantly outperforming the standard approaches and existing methods on different datasets.
Conclusions
The resulting accuracy of the proposed RL-based segmentation framework demonstrates an improvement against segmentation with standard U-Net architectures.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


