Overcoming data scarcity in biomedical imaging with a foundational multi-task model
IF 12
Q1 COMPUTER SCIENCE, INTERDISCIPLINARY APPLICATIONS
引用次数: 0
Abstract
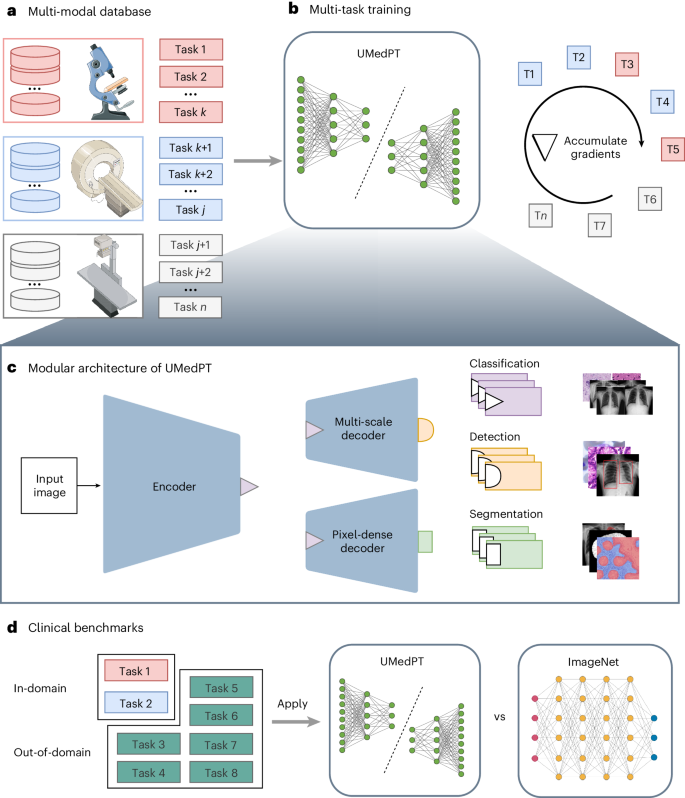
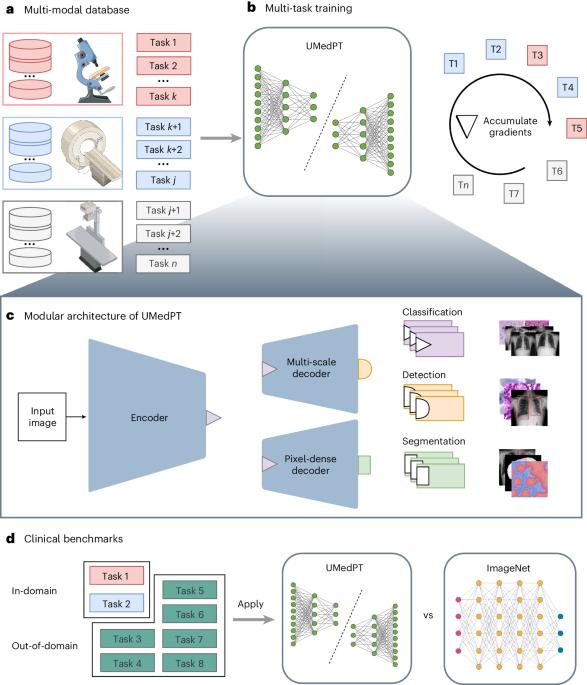
Foundational models, pretrained on a large scale, have demonstrated substantial success across non-medical domains. However, training these models typically requires large, comprehensive datasets, which contrasts with the smaller and more specialized datasets common in biomedical imaging. Here we propose a multi-task learning strategy that decouples the number of training tasks from memory requirements. We trained a universal biomedical pretrained model (UMedPT) on a multi-task database including tomographic, microscopic and X-ray images, with various labeling strategies such as classification, segmentation and object detection. The UMedPT foundational model outperformed ImageNet pretraining and previous state-of-the-art models. For classification tasks related to the pretraining database, it maintained its performance with only 1% of the original training data and without fine-tuning. For out-of-domain tasks it required only 50% of the original training data. In an external independent validation, imaging features extracted using UMedPT proved to set a new standard for cross-center transferability. UMedPT, a foundational model for biomedical imaging, has been trained on a variety of medical tasks with different types of label. It has achieved high performance with less training data in various clinical applications.
利用基础多任务模型克服生物医学成像中的数据匮乏问题。
经过大规模预训练的基础模型在非医疗领域取得了巨大成功。然而,训练这些模型通常需要大型、全面的数据集,这与生物医学成像中常见的更小、更专业的数据集形成了鲜明对比。在这里,我们提出了一种多任务学习策略,将训练任务的数量与内存要求分离开来。我们在一个多任务数据库上训练了一个通用生物医学预训练模型(UMedPT),该数据库包括断层扫描、显微镜和 X 射线图像,并采用了分类、分割和对象检测等多种标记策略。UMedPT 基础模型的表现优于 ImageNet 预训练模型和以前的先进模型。对于与预训练数据库相关的分类任务,只需使用 1%的原始训练数据,无需微调即可保持性能。对于域外任务,它只需要原始训练数据的 50%。在外部独立验证中,使用 UMedPT 提取的成像特征被证明是跨中心可转移性的新标准。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文

 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


